The evolution of transposable elements in natural populations of self-fertilizing Arabidopsis thaliana and its outcrossing relative Arabidopsis lyrata
- PMID: 20067644
- PMCID: PMC2837042
- DOI: 10.1186/1471-2148-10-10
The evolution of transposable elements in natural populations of self-fertilizing Arabidopsis thaliana and its outcrossing relative Arabidopsis lyrata
Abstract
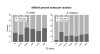
Background: Transposable Elements (TEs) make up the majority of plant genomes, and thus understanding TE evolutionary dynamics is key to understanding plant genome evolution. Plant reproductive systems are diverse and mating type variation is one factor among many hypothesized to influence TE evolutionary dynamics. Here, we collected a large TE-display data set in self-fertilizing Arabidopsis thaliana, and compared it to data gathered in outcrossing Arabidopsis lyrata. We analyzed seven TE families in four natural populations of each species to tease apart the effects of mating system, demography, transposition, and selection in determining patterns of TE diversity.
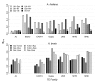
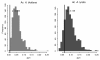
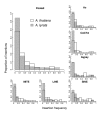
Results: Measures of TE band differentiation were largely consistent across TE families. However, patterns of diversity in A. thaliana Ac elements differed significantly from that other TEs, perhaps signaling a lack of recent transposition. Across TE families, we estimated higher allele frequencies and lower selection coefficients on A. thaliana TE insertions relative to A. lyrata TE insertions.
Conclusions: The differences in TE distributions between the two Arabidopsis species represents a synthesis of evolutionary forces that include the transposition dynamics of individual TE families and the demographic histories of populations. There are also species-specific differences that could be attributed to the effects of mating system, including higher overall allele frequencies in the selfing lineage and a greater proportion of among population TE diversity in the outcrossing lineage.
Figures
Similar articles
-
The predominantly selfing plant Arabidopsis thaliana experienced a recent reduction in transposable element abundance compared to its outcrossing relative Arabidopsis lyrata.Mob DNA. 2012 Feb 7;3(1):2. doi: 10.1186/1759-8753-3-2. Mob DNA. 2012. PMID: 22313744 Free PMC article.
-
Accumulation of transposable elements in selfing populations of Arabidopsis lyrata supports the ectopic recombination model of transposon evolution.New Phytol. 2018 Jul;219(2):767-778. doi: 10.1111/nph.15201. Epub 2018 May 14. New Phytol. 2018. PMID: 29757461
-
Demography and weak selection drive patterns of transposable element diversity in natural populations of Arabidopsis lyrata.Proc Natl Acad Sci U S A. 2008 Sep 16;105(37):13965-70. doi: 10.1073/pnas.0804671105. Epub 2008 Sep 4. Proc Natl Acad Sci U S A. 2008. PMID: 18772373 Free PMC article.
-
Mobility connects: transposable elements wire new transcriptional networks by transferring transcription factor binding motifs.Biochem Soc Trans. 2020 Jun 30;48(3):1005-1017. doi: 10.1042/BST20190937. Biochem Soc Trans. 2020. PMID: 32573687 Free PMC article. Review.
-
Transposable element evolution in plant genome ecosystems.Curr Opin Plant Biol. 2023 Oct;75:102418. doi: 10.1016/j.pbi.2023.102418. Epub 2023 Jul 15. Curr Opin Plant Biol. 2023. PMID: 37459733 Review.
Cited by
-
Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata.Proc Natl Acad Sci U S A. 2011 Feb 8;108(6):2322-7. doi: 10.1073/pnas.1018222108. Epub 2011 Jan 20. Proc Natl Acad Sci U S A. 2011. PMID: 21252301 Free PMC article.
-
Sizing up Arabidopsis genome evolution.Heredity (Edinb). 2011 Dec;107(6):509-10. doi: 10.1038/hdy.2011.47. Epub 2011 Jun 29. Heredity (Edinb). 2011. PMID: 21712843 Free PMC article. No abstract available.
-
Copy number variation in transcriptionally active regions of sexual and apomictic Boechera demonstrates independently derived apomictic lineages.Plant Cell. 2013 Oct;25(10):3808-23. doi: 10.1105/tpc.113.113860. Epub 2013 Oct 29. Plant Cell. 2013. PMID: 24170129 Free PMC article.
-
Co-evolution between transposable elements and their hosts: a major factor in genome size evolution?Chromosome Res. 2011 Aug;19(6):777-86. doi: 10.1007/s10577-011-9229-0. Chromosome Res. 2011. PMID: 21850458 Review.
-
Mating system shifts and transposable element evolution in the plant genus Capsella.BMC Genomics. 2014 Jul 16;15(1):602. doi: 10.1186/1471-2164-15-602. BMC Genomics. 2014. PMID: 25030755 Free PMC article.

