Population genetic inference from genomic sequence variation
- PMID: 20067940
- PMCID: PMC2840988
- DOI: 10.1101/gr.079509.108
Population genetic inference from genomic sequence variation
Abstract
Population genetics has evolved from a theory-driven field with little empirical data into a data-driven discipline in which genome-scale data sets test the limits of available models and computational analysis methods. In humans and a few model organisms, analyses of whole-genome sequence polymorphism data are currently under way. And in light of the falling costs of next-generation sequencing technologies, such studies will soon become common in many other organisms as well. Here, we assess the challenges to analyzing whole-genome sequence polymorphism data, and we discuss the potential of these data to yield new insights concerning population history and the genomic prevalence of natural selection.
Figures
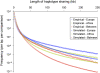

References
-
- Albrechtsen A, Sand Korneliussen T, Moltke I, van Overseem Hansen T, Nielsen FC, Nielsen R. Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium. Genet Epidemiol. 2009;33:266–274. - PubMed
-
- Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, Lander ES. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. - PubMed
-
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
