A single ClpS monomer is sufficient to direct the activity of the ClpA hexamer
- PMID: 20068042
- PMCID: PMC2838299
- DOI: 10.1074/jbc.M109.053736
A single ClpS monomer is sufficient to direct the activity of the ClpA hexamer
Abstract
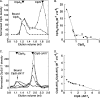
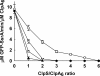
ClpS is an adaptor protein that interacts with ClpA and promotes degradation of proteins with N-end rule degradation motifs (N-degrons) by ClpAP while blocking degradation of substrates with other motifs. Although monomeric ClpS forms a 1:1 complex with an isolated N-domain of ClpA, only one molecule of ClpS binds with high affinity to ClpA hexamers (ClpA(6)). One or two additional molecules per hexamer bind with lower affinity. Tightly bound ClpS dissociates slowly from ClpA(6) with a t((1/2)) of approximately 3 min at 37 degrees C. Maximum activation of degradation of the N-end rule substrate, LR-GFP(Venus), occurs with a single ClpS bound per ClpA(6); one ClpS is also sufficient to inhibit degradation of proteins without N-degrons. ClpS competitively inhibits degradation of unfolded substrates that interact with ClpA N-domains and is a non-competitive inhibitor with substrates that depend on internal binding sites in ClpA. ClpS inhibition of substrate binding is dependent on the order of addition. When added first, ClpS blocks binding of both high and low affinity substrates; however, when substrates first form committed complexes with ClpA(6), ClpS cannot displace them or block their degradation by ClpP. We propose that the first molecule of ClpS binds to the N-domain and to an additional functional binding site, sterically blocking binding of non-N-end rule substrates as well as additional ClpS molecules to ClpA(6). Limiting ClpS-mediated substrate delivery to one per ClpA(6) avoids congestion at the axial channel and allows facile transfer of proteins to the unfolding and translocation apparatus.
Figures


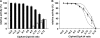

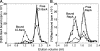

Similar articles
-
An intrinsic degradation tag on the ClpA C-terminus regulates the balance of ClpAP complexes with different substrate specificity.J Mol Biol. 2008 Dec 12;384(2):503-11. doi: 10.1016/j.jmb.2008.09.046. Epub 2008 Sep 26. J Mol Biol. 2008. PMID: 18835567
-
The molecular chaperone, ClpA, has a single high affinity peptide binding site per hexamer.J Biol Chem. 2005 Apr 1;280(13):12221-30. doi: 10.1074/jbc.M411733200. Epub 2005 Jan 18. J Biol Chem. 2005. PMID: 15657062
-
The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.Mol Cell. 2011 Jul 22;43(2):217-28. doi: 10.1016/j.molcel.2011.06.009. Mol Cell. 2011. PMID: 21777811 Free PMC article.
-
ClpP: a structurally dynamic protease regulated by AAA+ proteins.J Struct Biol. 2012 Aug;179(2):202-10. doi: 10.1016/j.jsb.2012.05.003. Epub 2012 May 14. J Struct Biol. 2012. PMID: 22595189 Review.
-
The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies.Trends Cell Biol. 2007 Apr;17(4):165-72. doi: 10.1016/j.tcb.2007.02.001. Epub 2007 Feb 15. Trends Cell Biol. 2007. PMID: 17306546 Review.
Cited by
-
The N-degradome of Escherichia coli: limited proteolysis in vivo generates a large pool of proteins bearing N-degrons.J Biol Chem. 2013 Oct 4;288(40):28913-24. doi: 10.1074/jbc.M113.492108. Epub 2013 Aug 19. J Biol Chem. 2013. PMID: 23960079 Free PMC article.
-
The Intrinsically Disordered N-terminal Extension of the ClpS Adaptor Reprograms Its Partner AAA+ ClpAP Protease.J Mol Biol. 2020 Aug 7;432(17):4908-4921. doi: 10.1016/j.jmb.2020.07.007. Epub 2020 Jul 17. J Mol Biol. 2020. PMID: 32687854 Free PMC article.
-
Structural Basis of an N-Degron Adaptor with More Stringent Specificity.Structure. 2016 Feb 2;24(2):232-42. doi: 10.1016/j.str.2015.12.008. Epub 2016 Jan 21. Structure. 2016. PMID: 26805523 Free PMC article.
-
Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion.J Biol Chem. 2011 Aug 26;286(34):30010-21. doi: 10.1074/jbc.M111.253377. Epub 2011 Jul 5. J Biol Chem. 2011. PMID: 21733841 Free PMC article.
-
Mycobacterium tuberculosis ClpC1 N-Terminal Domain Is Dispensable for Adaptor Protein-Dependent Allosteric Regulation.Int J Mol Sci. 2018 Nov 19;19(11):3651. doi: 10.3390/ijms19113651. Int J Mol Sci. 2018. PMID: 30463272 Free PMC article.
References
-
- Schirmer E. C., Glover J. R., Singer M. A., Lindquist S. (1996) Trends Biochem. Sci. 21, 289–296 - PubMed
-
- Gottesman S., Maurizi M. R., Wickner S. (1997) Cell 91, 435–438 - PubMed
-
- Levchenko I., Seidel M., Sauer R. T., Baker T. A. (2000) Science 289, 2354–2356 - PubMed
-
- Erbse A., Schmidt R., Bornemann T., Schneider-Mergener J., Mogk A., Zahn R., Dougan D. A., Bukau B. (2006) Nature 439, 753–756 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

