Gene expression profiling identifies Fibronectin 1 and CXCL9 as candidate biomarkers for breast cancer screening
- PMID: 20068563
- PMCID: PMC2822945
- DOI: 10.1038/sj.bjc.6605511
Gene expression profiling identifies Fibronectin 1 and CXCL9 as candidate biomarkers for breast cancer screening
Abstract
Background: There is a need to develop blood-based bioassays for breast cancer (BC) screening. In this study, differential gene expression between BC samples and benign tumours was used to identify candidate biomarkers for blood-based screening.
Methods: We identified two proteins (Fibronectin 1 and CXCL9) from a gene expression data set that included 120 BC samples and 45 benign lesions. These proteins fulfil the following criteria: differential gene expression between cancer and benign lesion, protein released in the extracellular medium and stable in the serum, commercially available ELISA kit, ELISA accuracy in a feasibility study. Protein concentrations were determined by ELISA. Blood samples were from normal volunteers (n=119) and early BC patients (n=133).
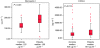
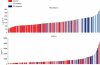
Results: Seventy-three per cent of patients had cT1-T2 tumour. Patients had higher CXCL9 and Fibronectin 1 concentrations than volunteers. CXCL9 mean concentration was 851 and 635 pg ml(-1) for patients and volunteers respectively (P=0.013). CXCL9 concentration was significantly higher in patients with estrogen receptor (ER)-negative compared with volunteers (P=0.003), data consistent with gene expression profile. Fibronectin 1 mean concentration was 190 microg ml(-1) for patients and 125 microg ml(-1) for volunteers (P<0.001). Areas under the curve for BC diagnosis were 0.78 and 0.62 for Fibronectin 1 and CXCL9 respectively. A combined score including Fibronectin 1 and CXCL9 dosages presented 53% of sensitivity and 98% of specificity. Similar performances were observed for ER-negative tumours.
Conclusions: This study suggests that Fibronectin 1/CXCL9 dosage in serum could screen a significant rate of BC, including ER-negative, and that differential gene expression analysis is a good approach to select candidate biomarkers to set up blood assays cancer screening.
Figures

Similar articles
-
Novel serum protein biomarker panel revealed by mass spectrometry and its prognostic value in breast cancer.Breast Cancer Res. 2014 Jun 16;16(3):R63. doi: 10.1186/bcr3676. Breast Cancer Res. 2014. PMID: 24935269 Free PMC article.
-
Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer.Oncotarget. 2016 Jun 28;7(26):40189-40199. doi: 10.18632/oncotarget.9561. Oncotarget. 2016. PMID: 27250024 Free PMC article.
-
Expression profiling identifies genes that predict recurrence of breast cancer after adjuvant CMF-based chemotherapy.Breast Cancer Res Treat. 2009 Nov;118(1):45-56. doi: 10.1007/s10549-008-0207-y. Epub 2008 Oct 17. Breast Cancer Res Treat. 2009. PMID: 18925433
-
The Role of CXCL13 and CXCL9 in Early Breast Cancer.Clin Breast Cancer. 2020 Feb;20(1):e36-e53. doi: 10.1016/j.clbc.2019.08.008. Epub 2019 Sep 5. Clin Breast Cancer. 2020. PMID: 31699671
-
Serum levels of chemokine (C-X-C motif) ligand 9 (CXCL9) are associated with tumor progression and treatment outcome in patients with oral cavity squamous cell carcinoma.Oral Oncol. 2013 Aug;49(8):802-7. doi: 10.1016/j.oraloncology.2013.05.006. Epub 2013 Jun 12. Oral Oncol. 2013. PMID: 23769451
Cited by
-
CXCL9 Is a Potential Biomarker of Immune Infiltration Associated With Favorable Prognosis in ER-Negative Breast Cancer.Front Oncol. 2021 Aug 30;11:710286. doi: 10.3389/fonc.2021.710286. eCollection 2021. Front Oncol. 2021. PMID: 34527583 Free PMC article.
-
Epoxy metabolites of linoleic acid promote the development of breast cancer via orchestrating PLEC/NFκB1/CXCL9-mediated tumor growth and metastasis.Cell Death Dis. 2024 Dec 18;15(12):901. doi: 10.1038/s41419-024-07300-6. Cell Death Dis. 2024. PMID: 39695149 Free PMC article.
-
Quantifying spatial CXCL9 distribution with image analysis predicts improved prognosis of triple-negative breast cancer.Front Genet. 2024 Jun 18;15:1421573. doi: 10.3389/fgene.2024.1421573. eCollection 2024. Front Genet. 2024. PMID: 38957805 Free PMC article.
-
SSL-VQ: vector-quantized variational autoencoders for semi-supervised prediction of therapeutic targets across diverse diseases.Bioinformatics. 2025 Feb 4;41(2):btaf039. doi: 10.1093/bioinformatics/btaf039. Bioinformatics. 2025. PMID: 39880378 Free PMC article.
-
MicroRNA-139 targets fibronectin 1 to inhibit papillary thyroid carcinoma progression.Oncol Lett. 2017 Dec;14(6):7799-7806. doi: 10.3892/ol.2017.7201. Epub 2017 Oct 17. Oncol Lett. 2017. Retraction in: Oncol Lett. 2022 May 31;24(1):237. doi: 10.3892/ol.2022.13357. PMID: 29250177 Free PMC article. Retracted.
References
-
- Andre F, Michiels S, Dessen P, Scott V, Suciu V, Uzan C, Lazar V, Lacroix L, Vassal G, Spielmann M, Vielh P, Delaloge S (2009) Exonic expression profile of breast tumors: a retrospective analysis of 165 samples from patients referred to a diagnosis center. Lancet Oncol 10(4): 381–390 - PubMed
-
- Bukowski RM, Rayman P, Molto L, Tannenbaum CS, Olencki T, Peereboom D, Tubbs R, McLain D, Budd GT, Griffin T, Novick A, Hamilton TA, Finke J (1999) Interferon-gamma and CXC chemokine induction by interleukin 12 in renal cell carcinoma. Clin Cancer Res 5(10): 2780–2789 - PubMed
-
- Couchman JR, Austria MR, Woods A (1990) Fibronectin–cell interaction. J Invest Dermatol 94: 7s–14s - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

