Detection and identification of old world Leishmania by high resolution melt analysis
- PMID: 20069036
- PMCID: PMC2797090
- DOI: 10.1371/journal.pntd.0000581
Detection and identification of old world Leishmania by high resolution melt analysis
Abstract
Background: Three major forms of human disease, cutaneous leishmaniasis, visceral leishmaniasis and mucocutaneous leishmaniasis, are caused by several leishmanial species whose geographic distribution frequently overlaps. These Leishmania species have diverse reservoir hosts, sand fly vectors and transmission patterns. In the Old World, the main parasite species responsible for leishmaniasis are Leishmania infantum, L. donovani, L. tropica, L. aethiopica and L. major. Accurate, rapid and sensitive diagnostic and identification procedures are crucial for the detection of infection and characterization of the causative leishmanial species, in order to provide accurate treatment, precise prognosis and appropriate public health control measures.
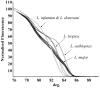
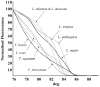
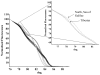
Methods/principal findings: High resolution melt analysis of a real time PCR product from the Internal Transcribed Spacer-1 rRNA region was used to identify and quantify Old World Leishmania in 300 samples from human patients, reservoir hosts and sand flies. Different characteristic high resolution melt analysis patterns were exhibited by L. major, L. tropica, L. aethiopica, and L. infantum. Genotyping by high resolution melt analysis was verified by DNA sequencing or restriction fragment length polymorphism. This new assay was able to detect as little as 2-4 ITS1 gene copies in a 5 microl DNA sample, i.e., less than a single parasite per reaction.
Conclusions/significance: This new technique is useful for rapid diagnosis of leishmaniasis and simultaneous identification and quantification of the infecting Leishmania species. It can be used for diagnostic purposes directly from clinical samples, as well as epidemiological studies, reservoir host investigations and vector surveys.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
A real-time ITS1-PCR based method in the diagnosis and species identification of Leishmania parasite from human and dog clinical samples in Turkey.PLoS Negl Trop Dis. 2013 May 9;7(5):e2205. doi: 10.1371/journal.pntd.0002205. Print 2013. PLoS Negl Trop Dis. 2013. PMID: 23675543 Free PMC article.
-
Molecular detection of Leishmania donovani, Leishmania major, and Trypanosoma species in Sergentomyia squamipleuris sand flies from a visceral leishmaniasis focus in Merti sub-County, eastern Kenya.Parasit Vectors. 2021 Jan 18;14(1):53. doi: 10.1186/s13071-020-04517-0. Parasit Vectors. 2021. PMID: 33461609 Free PMC article.
-
A High Resolution Melting Analysis (HRM) PCR assay for the detection and identification of Old World Leishmania species.PLoS Negl Trop Dis. 2024 Dec 23;18(12):e0012762. doi: 10.1371/journal.pntd.0012762. eCollection 2024 Dec. PLoS Negl Trop Dis. 2024. PMID: 39715284 Free PMC article.
-
The applicability of real-time PCR in the diagnostic of cutaneous leishmaniasis and parasite quantification for clinical management: Current status and perspectives.Acta Trop. 2018 Aug;184:29-37. doi: 10.1016/j.actatropica.2017.09.020. Epub 2017 Sep 29. Acta Trop. 2018. PMID: 28965842 Review.
-
Leishmaniasis in deployed military populations: A systematic review and meta-analysis.PLoS Negl Trop Dis. 2025 Mar 10;19(3):e0012680. doi: 10.1371/journal.pntd.0012680. eCollection 2025 Mar. PLoS Negl Trop Dis. 2025. PMID: 40063644 Free PMC article.
Cited by
-
Application of High-Resolution Melting (HRM) Technique towards the Detection of Asymptomatic Malaria in a Malaria Endemic Area of Southeastern Iran under Elimination Program.J Arthropod Borne Dis. 2020 Dec 31;14(4):353-362. doi: 10.18502/jad.v14i4.5272. eCollection 2020 Dec. J Arthropod Borne Dis. 2020. PMID: 33954209 Free PMC article.
-
Detection and molecular typing of Leishmania tropica from Phlebotomus sergenti and lesions of cutaneous leishmaniasis in an emerging focus of Morocco.Parasit Vectors. 2013 Jul 26;6:217. doi: 10.1186/1756-3305-6-217. Parasit Vectors. 2013. PMID: 23890256 Free PMC article.
-
Diagnosis and identification of Leishmania spp. from Giemsa-stained slides, by real-time PCR and melting curve analysis in south-west of Iran.Ann Trop Med Parasitol. 2011 Dec;105(8):559-65. doi: 10.1179/2047773211Y.0000000014. Ann Trop Med Parasitol. 2011. PMID: 22325815 Free PMC article.
-
HRM Accuracy and Limitations as a Species Typing Tool for Leishmania Parasites.Int J Mol Sci. 2023 Sep 30;24(19):14784. doi: 10.3390/ijms241914784. Int J Mol Sci. 2023. PMID: 37834232 Free PMC article.
-
Canine leishmaniosis caused by Leishmania major and Leishmania tropica: comparative findings and serology.Parasit Vectors. 2017 Mar 13;10(1):113. doi: 10.1186/s13071-017-2050-7. Parasit Vectors. 2017. PMID: 28285601 Free PMC article.
References
-
- Schönian G, Mauricio I, Gramiccia M, Cañavate C, Boelaert M, et al. Leishmaniases in the Mediterranean in the era of molecular epidemiology. Trends Parasitol. 2008;24:135–142. - PubMed
-
- Bañuls AL, Guerrini F, Le Pont F, Barrera C, Espinel I, et al. Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J Eukaryot Microbiol. 1997;44:408–4011. - PubMed
-
- Mauricio IL, Stothard JR, Miles MA. Leishmania donovani complex: genotyping with the ribosomal internal transcribed spacer and the mini-exon. Parasitology. 2004;28:263–267. - PubMed
-
- Rioux JA, Lanotte G, Serres E, Pratlong E, Bastien P, et al. Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann Parasitol Hum Comp. 1990;65:111–125. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

