Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts
- PMID: 20070886
- PMCID: PMC2847717
- DOI: 10.1186/gb-2010-11-1-r5
Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts
Abstract
Background: Radial chromosome positioning in interphase nuclei is nonrandom and can alter according to developmental, differentiation, proliferation, or disease status. However, it is not yet clear when and how chromosome repositioning is elicited.
Results: By investigating the positioning of all human chromosomes in primary fibroblasts that have left the proliferative cell cycle, we have demonstrated that in cells made quiescent by reversible growth arrest, chromosome positioning is altered considerably. We found that with the removal of serum from the culture medium, chromosome repositioning took less than 15 minutes, required energy and was inhibited by drugs affecting the polymerization of myosin and actin. We also observed that when cells became quiescent, the nuclear distribution of nuclear myosin 1 beta was dramatically different from that in proliferating cells. If we suppressed the expression of nuclear myosin 1 beta by using RNA-interference procedures, the movement of chromosomes after 15 minutes in low serum was inhibited. When high serum was restored to the serum-starved cultures, chromosome repositioning was evident only after 24 to 36 hours, and this coincided with a return to a proliferating distribution of nuclear myosin 1 beta.
Conclusions: These findings demonstrate that genome organization in interphase nuclei is altered considerably when cells leave the proliferative cell cycle and that repositioning of chromosomes relies on efficient functioning of an active nuclear motor complex that contains nuclear myosin 1 beta.
Figures
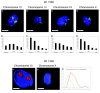
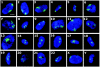

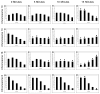
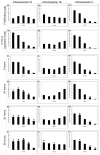



Comment in
-
Chromosome territory relocation paradigm during DNA damage response: Some insights from molecular biology to physics.Nucleus. 2017 Sep 3;8(5):449-460. doi: 10.1080/19491034.2017.1313938. Epub 2017 Jun 22. Nucleus. 2017. PMID: 28640660 Free PMC article.
Similar articles
-
Interphase Chromosomes in Replicative Senescence: Chromosome Positioning as a Senescence Biomarker and the Lack of Nuclear Motor-Driven Chromosome Repositioning in Senescent Cells.Front Cell Dev Biol. 2021 May 24;9:640200. doi: 10.3389/fcell.2021.640200. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34113611 Free PMC article.
-
Chromobility: the rapid movement of chromosomes in interphase nuclei.Biochem Soc Trans. 2011 Dec;39(6):1747-51. doi: 10.1042/BST20110696. Biochem Soc Trans. 2011. PMID: 22103519 Review.
-
Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis.Mol Biol Cell. 2004 Jan;15(1):197-206. doi: 10.1091/mbc.e03-06-0388. Epub 2003 Nov 14. Mol Biol Cell. 2004. PMID: 14617810 Free PMC article.
-
Genome organization: Tag it, move it, place it.Curr Opin Cell Biol. 2021 Feb;68:90-97. doi: 10.1016/j.ceb.2020.10.005. Epub 2020 Nov 7. Curr Opin Cell Biol. 2021. PMID: 33166737 Free PMC article. Review.
-
Chromosome territory relocation paradigm during DNA damage response: Some insights from molecular biology to physics.Nucleus. 2017 Sep 3;8(5):449-460. doi: 10.1080/19491034.2017.1313938. Epub 2017 Jun 22. Nucleus. 2017. PMID: 28640660 Free PMC article.
Cited by
-
The nuclear envelope at a glance.J Cell Sci. 2010 Jun 15;123(Pt 12):1973-8. doi: 10.1242/jcs.019042. J Cell Sci. 2010. PMID: 20519579 Free PMC article. No abstract available.
-
Cytochalasin D restores nuclear size acting on F-actin and IZUMO1 localization in low-quality spermatozoa.Int J Biol Sci. 2023 Apr 17;19(7):2234-2255. doi: 10.7150/ijbs.77166. eCollection 2023. Int J Biol Sci. 2023. PMID: 37151878 Free PMC article.
-
A meeting at risk: Unrepaired DSBs go for broke.Nucleus. 2017 Nov 2;8(6):589-599. doi: 10.1080/19491034.2017.1380138. Epub 2017 Nov 17. Nucleus. 2017. PMID: 29099269 Free PMC article. Review.
-
New Insights into Cellular Functions of Nuclear Actin.Biology (Basel). 2021 Apr 7;10(4):304. doi: 10.3390/biology10040304. Biology (Basel). 2021. PMID: 33916969 Free PMC article. Review.
-
Nuclear myosin 1 contributes to a chromatin landscape compatible with RNA polymerase II transcription activation.BMC Biol. 2015 Jun 5;13:35. doi: 10.1186/s12915-015-0147-z. BMC Biol. 2015. PMID: 26044184 Free PMC article.
References
-
- Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective; part II: fall and resurrection of chromosome territories during the 1950s to 1980s; part III: chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur J Histochem. 2006;50:223–272. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

