Distinct transcriptomes define rostral and caudal serotonin neurons
- PMID: 20071532
- PMCID: PMC3403750
- DOI: 10.1523/JNEUROSCI.4656-09.2010
Distinct transcriptomes define rostral and caudal serotonin neurons
Abstract
The molecular architecture of developing serotonin (5HT) neurons is poorly understood, yet its determination is likely to be essential for elucidating functional heterogeneity of these cells and the contribution of serotonergic dysfunction to disease pathogenesis. Here, we describe the purification of postmitotic embryonic 5HT neurons by flow cytometry for whole-genome microarray expression profiling of this unitary monoaminergic neuron type. Our studies identified significantly enriched expression of hundreds of unique genes in 5HT neurons, thus providing an abundance of new serotonergic markers. Furthermore, we identified several hundred transcripts encoding homeodomain, axon guidance, cell adhesion, intracellular signaling, ion transport, and imprinted genes associated with various neurodevelopmental disorders that were differentially enriched in developing rostral and caudal 5HT neurons. These findings suggested a homeodomain code that distinguishes rostral and caudal 5HT neurons. Indeed, verification studies demonstrated that Hmx homeodomain and Hox gene expression defined an Hmx(+) rostral subtype and Hox(+) caudal subtype. Expression of engrailed genes in a subset of 5HT neurons in the rostral domain further distinguished two subtypes defined as Hmx(+)En(+) and Hmx(+)En(-). The differential enrichment of gene sets for different canonical pathways and gene ontology categories provided additional evidence for heterogeneity between rostral and caudal 5HT neurons. These findings demonstrate a deep transcriptome and biological pathway duality for neurons that give rise to the ascending and descending serotonergic subsystems. Our databases provide a rich, clinically relevant resource for definition of 5HT neuron subtypes and elucidation of the genetic networks required for serotonergic function.
Figures
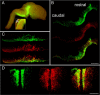
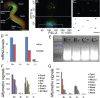

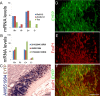
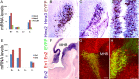

Similar articles
-
A genetic approach to access serotonin neurons for in vivo and in vitro studies.Proc Natl Acad Sci U S A. 2005 Nov 8;102(45):16472-7. doi: 10.1073/pnas.0504510102. Epub 2005 Oct 26. Proc Natl Acad Sci U S A. 2005. PMID: 16251278 Free PMC article.
-
Hedgehog and Fgf signaling pathways regulate the development of tphR-expressing serotonergic raphe neurons in zebrafish embryos.J Neurobiol. 2004 Sep 5;60(3):275-88. doi: 10.1002/neu.20023. J Neurobiol. 2004. PMID: 15281067 Free PMC article.
-
Molecular genetics of the early development of hindbrain serotonergic neurons.Clin Genet. 2005 Dec;68(6):487-94. doi: 10.1111/j.1399-0004.2005.00534.x. Clin Genet. 2005. PMID: 16283875 Review.
-
Location and size of dopaminergic and serotonergic cell populations are controlled by the position of the midbrain-hindbrain organizer.J Neurosci. 2003 May 15;23(10):4199-207. doi: 10.1523/JNEUROSCI.23-10-04199.2003. J Neurosci. 2003. PMID: 12764108 Free PMC article.
-
Redefining the serotonergic system by genetic lineage.Nat Neurosci. 2008 Apr;11(4):417-9. doi: 10.1038/nn2050. Epub 2008 Mar 16. Nat Neurosci. 2008. PMID: 18344997 Free PMC article.
Cited by
-
Serotonin neuron diversity in the dorsal raphe.ACS Chem Neurosci. 2013 Jan 16;4(1):22-5. doi: 10.1021/cn300224n. ACS Chem Neurosci. 2013. PMID: 23336040 Free PMC article. Review.
-
Probing the diversity of serotonin neurons.Philos Trans R Soc Lond B Biol Sci. 2012 Sep 5;367(1601):2382-94. doi: 10.1098/rstb.2011.0378. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22826339 Free PMC article. Review.
-
Central respiratory chemoreception.J Comp Neurol. 2010 Oct 1;518(19):3883-906. doi: 10.1002/cne.22435. J Comp Neurol. 2010. PMID: 20737591 Free PMC article. Review.
-
Embracing diversity in the 5-HT neuronal system.Nat Rev Neurosci. 2019 Jul;20(7):397-424. doi: 10.1038/s41583-019-0151-3. Nat Rev Neurosci. 2019. PMID: 30948838 Review.
-
5-HT1A Receptor-Mediated Autoinhibition and the Control of Serotonergic Cell Firing.ACS Chem Neurosci. 2015 Jul 15;6(7):1110-5. doi: 10.1021/acschemneuro.5b00034. Epub 2015 May 26. ACS Chem Neurosci. 2015. PMID: 25913021 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
