Identification of novel diagnostic serum biomarkers for Chagas' disease in asymptomatic subjects by mass spectrometric profiling
- PMID: 20071547
- PMCID: PMC2849606
- DOI: 10.1128/JCM.02207-09
Identification of novel diagnostic serum biomarkers for Chagas' disease in asymptomatic subjects by mass spectrometric profiling
Abstract
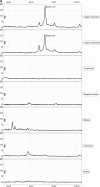
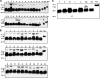
More than 10 million people are thought to be infected with Trypanosoma cruzi, primarily in the Americas. The clinical manifestations of Chagas' disease (CD) are variable, but most subjects remain asymptomatic for decades. Only 15 to 30% eventually develop terminal complications. All current diagnostic tests have limitations. New approaches are needed for blood bank screening as well as for improved diagnosis and prognosis. Sera from subjects with asymptomatic CD (n = 131) were compared to those from uninfected controls (n = 164) and subjects with other parasitic diseases (n = 140), using protein array mass spectrometry. To identify biomarkers associated with CD, sera were fractionated by anion-exchange chromatography and bound to two commercial ProteinChip array chemistries: WCX2 and IMAC3. Multiple candidate biomarkers were found in CD sera (3 to 75.4 kDa). Algorithms employing 3 to 5 of these biomarkers achieved up to 100% sensitivity and 98% specificity for CD. The biomarkers most useful for diagnosis were identified and validated. These included MIP1 alpha, C3a anaphylatoxin, and unusually truncated forms of fibronectin, apolipoprotein A1 (ApoA1), and C3. An antipeptide antiserum against the 28.9-kDa C terminus of the fibronectin fragment achieved good specificity (90%) for CD in a Western blot format. We identified full-length ApoA1 (28.1 kDa), the major structural and functional protein component of high-density lipoprotein (HDL), as an important negative biomarker for CD, and relatively little full-length ApoA1 was detected in CD sera. This work provides proof of principle that both platform-dependent (i.e., mass spectrometry-based) and platform-independent (i.e., Western blot) tests can be generated using high-throughput mass profiling.
Figures


Similar articles
-
85-kDa protein of Trypanosoma cruzi purified by affinity chromatography used in the multiple antigen binding assay (MABA) for the diagnosis of T. cruzi infection in a Venezuelan rural community.Parasitol Res. 2010 Apr;106(5):1127-34. doi: 10.1007/s00436-010-1773-6. Epub 2010 Feb 24. Parasitol Res. 2010. PMID: 20180133
-
Characterization of an immunodominant antigenic epitope from Trypanosoma cruzi as a biomarker of chronic Chagas' disease pathology.Clin Vaccine Immunol. 2012 Feb;19(2):167-73. doi: 10.1128/CVI.05566-11. Epub 2011 Dec 7. Clin Vaccine Immunol. 2012. PMID: 22155766 Free PMC article.
-
Apolipoprotein A1 and Fibronectin Fragments as Markers of Cure for the Chagas Disease.Methods Mol Biol. 2019;1955:263-273. doi: 10.1007/978-1-4939-9148-8_20. Methods Mol Biol. 2019. PMID: 30868534
-
Purified excreted-secreted antigens from Trypanosoma cruzi trypomastigotes as tools for diagnosis of Chagas' disease.J Clin Microbiol. 2006 Feb;44(2):291-6. doi: 10.1128/JCM.44.2.291-296.2006. J Clin Microbiol. 2006. PMID: 16455872 Free PMC article.
-
Strategies for prevention of transfusion-associated Chagas' disease.Transfus Med Rev. 1996 Jul;10(3):161-70. doi: 10.1016/s0887-7963(96)80057-5. Transfus Med Rev. 1996. PMID: 8809967 Review.
Cited by
-
Protective effect of the Japanese traditional medicine juzentaihoto on myelosuppression induced by the anticancer drug TS-1 and identification of a potential biomarker of this effect.BMC Complement Altern Med. 2012 Aug 9;12:118. doi: 10.1186/1472-6882-12-118. BMC Complement Altern Med. 2012. PMID: 22876791 Free PMC article.
-
A family cluster of Chagas disease detected through selective screening of blood donors: A case report and brief review.Can J Infect Dis Med Microbiol. 2015 May-Jun;26(3):157-61. doi: 10.1155/2015/628981. Can J Infect Dis Med Microbiol. 2015. PMID: 26236358 Free PMC article.
-
Cardiovascular biomarkers as predictors of adverse outcomes in chronic Chagas cardiomyopathy.PLoS One. 2021 Oct 28;16(10):e0258622. doi: 10.1371/journal.pone.0258622. eCollection 2021. PLoS One. 2021. PMID: 34710112 Free PMC article.
-
Proteomic identification of plasma protein tyrosine phosphatase alpha and fibronectin associated with liver fluke, Opisthorchis viverrini, infection.PLoS One. 2012;7(9):e45460. doi: 10.1371/journal.pone.0045460. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23029023 Free PMC article.
-
Role of the Complement System in the Modulation of T-Cell Responses in Chronic Chagas Disease.Front Cell Infect Microbiol. 2022 Jun 30;12:910854. doi: 10.3389/fcimb.2022.910854. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35846776 Free PMC article. Review.
References
-
- Antunes, C. M. F. 1999. Chagas disease (American trypanosomiasis): the epidemiology of Chagas disease, p. 351-369. In H. M. Gilles (ed.), Protozoal diseases. Arnold, London, United Kingdom.
-
- Atwood, J. A., III, D. B. Weatherly, T. A. Minning, B. Bundy, C. Cavola, F. R. Opperdoes, R. Orlando, and R. L. Tarleton. 2005. The Trypanosoma cruzi proteome. Science 309:473-476. - PubMed
-
- Baral, T. N., S. Magez, B. Stijlemans, K. Conrath, B. Vanhollebeke, E. Pays, S. Muyldermans, and P. De Baetselier. 2006. Experimental therapy of African trypanosomiasis with a nanobody-conjugated human trypanolytic factor. Nat. Med. 12:580-584. - PubMed
-
- Berasain, P., C. Carmona, B. Frangione, J. J. Cazzulo, and F. Goni. 2003. Specific cleavage sites on human IgG subclasses by cruzipain, the major cysteine proteinase from Trypanosoma cruzi. Mol. Biochem. Parasitol. 130:23-29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

