Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus
- PMID: 20071564
- PMCID: PMC2838145
- DOI: 10.1128/JVI.02226-09
Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus
Abstract
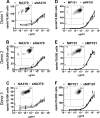
Very limited evidence has been reported to show human adaptive immune responses to the 2009 pandemic H1N1 swine-origin influenza A virus (S-OIV). We studied 17 S-OIV peptides homologous to immunodominant CD4 T epitopes from hemagglutinin (HA), neuraminidase (NA), nuclear protein (NP), M1 matrix protein (MP), and PB1 of a seasonal H1N1 strain. We concluded that 15 of these 17 S-OIV peptides would induce responses of seasonal influenza virus-specific T cells. Of these, seven S-OIV sequences were identical to seasonal influenza virus sequences, while eight had at least one amino acid that was not conserved. T cells recognizing epitopes derived from these S-OIV antigens could be detected ex vivo. Most of these T cells expressed memory markers, although none of the donors had been exposed to S-OIV. Functional analysis revealed that specific amino acid differences in the sequences of these S-OIV peptides would not affect or partially affect memory T-cell responses. These findings suggest that without protective antibody responses, individuals vaccinated against seasonal influenza A may still benefit from preexisting cross-reactive memory CD4 T cells reducing their susceptibility to S-OIV infection.
Figures

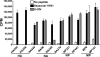



References
-
- CDC. 2009. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb. Mortal. Wkly. Rep. 58:521-524. - PubMed
-
- Dawood, F. S., S. Jain, L. Finelli, M. W. Shaw, S. Lindstrom, R. J. Garten, L. V. Gubareva, X. Xu, C. B. Bridges, and T. M. Uyeki. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605-2615. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

