Poly(A) at the 3' end of positive-strand RNA and VPg-linked poly(U) at the 5' end of negative-strand RNA are reciprocal templates during replication of poliovirus RNA
- PMID: 20071574
- PMCID: PMC2826026
- DOI: 10.1128/JVI.02620-08
Poly(A) at the 3' end of positive-strand RNA and VPg-linked poly(U) at the 5' end of negative-strand RNA are reciprocal templates during replication of poliovirus RNA
Abstract
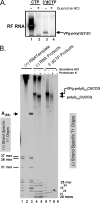
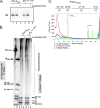
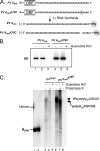
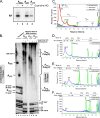
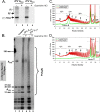
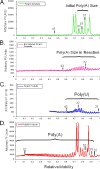
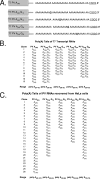
A 3' poly(A) tail is a common feature of picornavirus RNA genomes and the RNA genomes of many other positive-strand RNA viruses. We examined the manner in which the homopolymeric poly(A) and poly(U) portions of poliovirus (PV) positive- and negative-strand RNAs were used as reciprocal templates during RNA replication. Poly(A) sequences at the 3' end of viral positive-strand RNA were transcribed into VPg-linked poly(U) products at the 5' end of negative-strand RNA during PV RNA replication. Subsequently, VPg-linked poly(U) sequences at the 5' ends of negative-strand RNA templates were transcribed into poly(A) sequences at the 3' ends of positive-strand RNAs. The homopolymeric poly(A) and poly(U) portions of PV RNA products of replication were heterogeneous in length and frequently longer than the corresponding homopolymeric sequences of the respective viral RNA templates. The data support a model of PV RNA replication wherein reiterative transcription of homopolymeric templates ensures the synthesis of long 3' poly(A) tails on progeny RNA genomes.
Figures


References
-
- Baltimore, D., Y. Becker, and J. E. Darnell. 1964. Virus-specific double-stranded RNA in poliovirus-infected cells. Science 143:1034-1036. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

