Self versus non-self discrimination during CRISPR RNA-directed immunity
- PMID: 20072129
- PMCID: PMC2813891
- DOI: 10.1038/nature08703
Self versus non-self discrimination during CRISPR RNA-directed immunity
Abstract
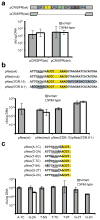
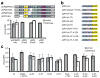
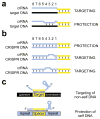
All immune systems must distinguish self from non-self to repel invaders without inducing autoimmunity. Clustered, regularly interspaced, short palindromic repeat (CRISPR) loci protect bacteria and archaea from invasion by phage and plasmid DNA through a genetic interference pathway. CRISPR loci are present in approximately 40% and approximately 90% of sequenced bacterial and archaeal genomes, respectively, and evolve rapidly, acquiring new spacer sequences to adapt to highly dynamic viral populations. Immunity requires a sequence match between the invasive DNA and the spacers that lie between CRISPR repeats. Each cluster is genetically linked to a subset of the cas (CRISPR-associated) genes that collectively encode >40 families of proteins involved in adaptation and interference. CRISPR loci encode small CRISPR RNAs (crRNAs) that contain a full spacer flanked by partial repeat sequences. CrRNA spacers are thought to identify targets by direct Watson-Crick pairing with invasive 'protospacer' DNA, but how they avoid targeting the spacer DNA within the encoding CRISPR locus itself is unknown. Here we have defined the mechanism of CRISPR self/non-self discrimination. In Staphylococcus epidermidis, target/crRNA mismatches at specific positions outside of the spacer sequence license foreign DNA for interference, whereas extended pairing between crRNA and CRISPR DNA repeats prevents autoimmunity. Hence, this CRISPR system uses the base-pairing potential of crRNAs not only to specify a target, but also to spare the bacterial chromosome from interference. Differential complementarity outside of the spacer sequence is a built-in feature of all CRISPR systems, indicating that this mechanism is a broadly applicable solution to the self/non-self dilemma that confronts all immune pathways.
Figures

References
-
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
-
- Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. - PubMed
-
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

