Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway
- PMID: 20072604
- PMCID: PMC2795201
- DOI: 10.1371/journal.ppat.1000712
Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway
Abstract
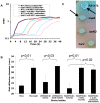
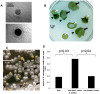
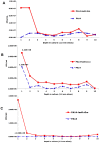
Microbes are subjected to selective pressures during chronic infections of host tissues. Pseudomonas aeruginosa isolates with inactivating mutations in the transcriptional regulator LasR are frequently selected within the airways of people with cystic fibrosis (CF), and infection with these isolates has been associated with poorer lung function outcomes. The mechanisms underlying selection for lasR mutation are unknown but have been postulated to involve the abundance of specific nutrients within CF airway secretions. We characterized lasR mutant P. aeruginosa strains and isolates to identify conditions found in CF airways that select for growth of lasR mutants. Relative to wild-type P. aeruginosa, lasR mutants exhibited a dramatic metabolic shift, including decreased oxygen consumption and increased nitrate utilization, that is predicted to confer increased fitness within the nutrient conditions known to occur in CF airways. This metabolic shift exhibited by lasR mutants conferred resistance to two antibiotics used frequently in CF care, tobramycin and ciprofloxacin, even under oxygen-dependent growth conditions, yet selection for these mutants in vitro did not require preceding antibiotic exposure. The selection for loss of LasR function in vivo, and the associated adverse clinical impact, could be due to increased bacterial growth in the oxygen-poor and nitrate-rich CF airway, and from the resulting resistance to therapeutic antibiotics. The metabolic similarities among diverse chronic infection-adapted bacteria suggest a common mode of adaptation and antibiotic resistance during chronic infection that is primarily driven by bacterial metabolic shifts in response to nutrient availability within host tissues.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

