Combining structure and sequence information allows automated prediction of substrate specificities within enzyme families
- PMID: 20072606
- PMCID: PMC2796266
- DOI: 10.1371/journal.pcbi.1000636
Combining structure and sequence information allows automated prediction of substrate specificities within enzyme families
Abstract
An important aspect of the functional annotation of enzymes is not only the type of reaction catalysed by an enzyme, but also the substrate specificity, which can vary widely within the same family. In many cases, prediction of family membership and even substrate specificity is possible from enzyme sequence alone, using a nearest neighbour classification rule. However, the combination of structural information and sequence information can improve the interpretability and accuracy of predictive models. The method presented here, Active Site Classification (ASC), automatically extracts the residues lining the active site from one representative three-dimensional structure and the corresponding residues from sequences of other members of the family. From a set of representatives with known substrate specificity, a Support Vector Machine (SVM) can then learn a model of substrate specificity. Applied to a sequence of unknown specificity, the SVM can then predict the most likely substrate. The models can also be analysed to reveal the underlying structural reasons determining substrate specificities and thus yield valuable insights into mechanisms of enzyme specificity. We illustrate the high prediction accuracy achieved on two benchmark data sets and the structural insights gained from ASC by a detailed analysis of the family of decarboxylating dehydrogenases. The ASC web service is available at http://asc.informatik.uni-tuebingen.de/.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

 . Alternatively, kernels may be used. (E) The final ASC model is trained using the generated feature vectors.
. Alternatively, kernels may be used. (E) The final ASC model is trained using the generated feature vectors.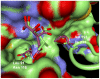
References
-
- Valencia A. Automatic annotation of protein function. Curr Opin Struct Biol. 2005;15:267–274. - PubMed
-
- Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press; 1998.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

