Hydrogen bonding progressively strengthens upon transfer of the protein urea-denatured state to water and protecting osmolytes
- PMID: 20073511
- PMCID: PMC2817916
- DOI: 10.1021/bi9015499
Hydrogen bonding progressively strengthens upon transfer of the protein urea-denatured state to water and protecting osmolytes
Abstract
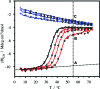
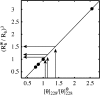
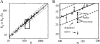
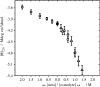
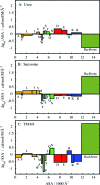
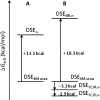
Using osmolyte cosolvents, we show that hydrogen-bonding contributions can be separated from hydrophobic interactions in the denatured state ensemble (DSE). Specifically, the effects of urea and the protecting osmolytes sarcosine and TMAO are reported on the thermally unfolded DSE of Nank4-7*, a truncated notch ankyrin protein. The high thermal energy of this state in the presence and absence of 6 M urea or 1 M sarcosine solution is sufficient to allow large changes in the hydrodynamic radius (R(h)) and secondary structure accretion without populating the native state. The CD change at 228 nm is proportional to the inverse of the volume of the DSE, giving a compact species equivalent to a premolten globule in 1 M sarcosine. The same general effects portraying hierarchical folding observed in the DSE at 55 degrees C are also often seen at room temperature. Analysis of Nank4-7* DSE structural energetics at room temperature as a function of solvent provides rationale for understanding the structural and dimensional effects in terms of how modulation of the solvent alters solvent quality for the peptide backbone. Results show that while the strength of hydrophobic interactions changes little on transferring the DSE from 6 M urea to water and then to 1 M TMAO, backbone-backbone (hydrogen-bonding) interactions are greatly enhanced due to progressively poorer solvent quality for the peptide backbone. Thus, increased intrachain hydrogen bonding guides secondary structure accretion and DSE contraction as solvent quality is decreased. This process is accompanied by increasing hydrophobic contacts as chain contraction gathers hydrophobes into proximity and the declining urea-backbone free energy gradient reaches urea concentrations that are energetically insufficient to keep hydrophobes apart in the DSE.
Figures
Similar articles
-
Hydrophobic Interactions in Aqueous Osmolyte Solutions: Thermodynamics of Solvation and Implication on Protein Stability.J Phys Chem B. 2025 May 29;129(21):5150-5165. doi: 10.1021/acs.jpcb.5c00785. Epub 2025 May 19. J Phys Chem B. 2025. PMID: 40387874 Free PMC article.
-
Structure and energetics of the hydrogen-bonded backbone in protein folding.Annu Rev Biochem. 2008;77:339-62. doi: 10.1146/annurev.biochem.77.061306.131357. Annu Rev Biochem. 2008. PMID: 18518824 Review.
-
Structure and effect of sarcosine on water and urea by using molecular dynamics simulations: Implications in protein stabilization.Biophys Chem. 2013 Jan;171:9-15. doi: 10.1016/j.bpc.2012.11.004. Epub 2012 Dec 4. Biophys Chem. 2013. PMID: 23266436
-
Interactions of S-peptide analogue in aqueous urea and trimethylamine-N-oxide solutions: a molecular dynamics simulation study.J Chem Phys. 2013 Jul 21;139(3):034504. doi: 10.1063/1.4813502. J Chem Phys. 2013. PMID: 23883044
-
Cosolvent effects on protein stability.Annu Rev Phys Chem. 2013;64:273-93. doi: 10.1146/annurev-physchem-040412-110156. Epub 2013 Jan 4. Annu Rev Phys Chem. 2013. PMID: 23298246 Review.
Cited by
-
Counting peptide-water hydrogen bonds in unfolded proteins.Protein Sci. 2011 Feb;20(2):417-27. doi: 10.1002/pro.574. Protein Sci. 2011. PMID: 21280132 Free PMC article.
-
Untangling a Structurally Resolved Protein Folding Intermediate.Biophys J. 2016 Mar 29;110(6):1205-6. doi: 10.1016/j.bpj.2016.01.028. Biophys J. 2016. PMID: 27028629 Free PMC article. No abstract available.
-
Phenylalanine as an effective stabilizer and aggregation inhibitor of Bacillus amyloliquefaciens alpha-amylase.AMB Express. 2024 Jun 8;14(1):69. doi: 10.1186/s13568-024-01712-5. AMB Express. 2024. PMID: 38850460 Free PMC article.
-
Beneficial effect of sugar osmolytes on the refolding of guanidine hydrochloride-denatured trehalose-6-phosphate hydrolase from Bacillus licheniformis.Biomed Res Int. 2015;2015:806847. doi: 10.1155/2015/806847. Epub 2015 Jan 13. Biomed Res Int. 2015. PMID: 25667926 Free PMC article.
-
Probing the Action of Chemical Denaturant on an Intrinsically Disordered Protein by Simulation and Experiment.J Am Chem Soc. 2016 Sep 14;138(36):11702-13. doi: 10.1021/jacs.6b05443. Epub 2016 Sep 1. J Am Chem Soc. 2016. PMID: 27583687 Free PMC article.
References
-
- Bolen D. W.; Rose G. D. (2008) Structure and energetics of the hydrogen-bonded backbone in protein folding. Annu. Rev. Biochem. 77, 339–362. - PubMed
-
- Hochachka P. W., and Somero G. N. (2002) Biochemical Adaptation. Mechanism and Process in Physiological Evolution, Oxford University Press, Oxford.
-
- Yancey P. H.; Clark M. E.; Hand S. C.; Bowlus R. D.; Somero G. N. (1982) Living with water stress: evolution of osmolyte systems. Science 217, 1214–1222. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

