Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens
- PMID: 20074338
- PMCID: PMC2820457
- DOI: 10.1186/1471-2164-11-29
Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens
Abstract
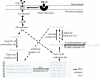
Background: The chemical components of sex pheromones have been determined for more than a thousand moth species, but so far only a handful of genes encoding enzymes responsible for the biosynthesis of these compounds have been identified. For understanding the evolution of moth sexual communication, it is essential to know which genes are involved in the production of specific pheromone components and what controls the variation in their relative frequencies in the pheromone blend. We used a transcriptomic approach to characterize the pheromone gland of the Noctuid moth Heliothis virescens, an important agricultural pest, in order to obtain substantial general sequence information and to identify a range of candidate genes involved in the pheromone biosynthetic pathway.
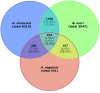
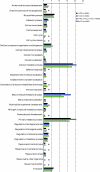
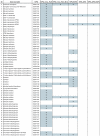
Results: To facilitate identifying sets of genes involved in a broad range of processes and to capture rare transcripts, we developed our majority of ESTs from a normalized cDNA library of Heliothis virescens pheromone glands (PG). Combining these with a non-normalized library yielded a total of 17,233 ESTs, which assembled into 2,082 contigs and 6,228 singletons. Using BLAST searches of the NR and Swissprot databases we were able to identify a large number of putative unique gene elements (unigenes), which we compared to those derived from previous transcriptomic surveys of the larval stage of Heliothis virescens. The distribution of unigenes among GO Biological Process functional groups shows an overall similarity between PG and larval transcriptomes, but with distinct enrichment of specific pathways in the PG. In addition, we identified a large number of candidate genes in the pheromone biosynthetic pathways.
Conclusion: These data constitute one of the first large-scale EST-projects for Noctuidae, a much-needed resource for exploring these pest species. Our analysis shows a surprisingly complex transcriptome and we identified a large number of potential pheromone biosynthetic pathway and immune-related genes that can be applied to population and systematic studies of Heliothis virescens and other Noctuidae.
Figures





References
-
- Butlin R, Trickett AJ. In: Insect pheromone research: New directions. Carde RT, Minks AK, editor. New York: Chapman and Hall; 1997. Can population genetic simulations help to interpret pheromone evolution? pp. 548–562.
-
- Cardé RT, Haynes KF. In: Advances in insect chemical ecology. Carde RT, Millar JG, editor. Cambridge, UK: Cambridge University Press; 2004. Structure of the pheromone communication channel in moths; pp. 283–332. full_text.
-
- Löfstedt C. Moth pheromone genetics and evolution. Philos Trans R Soc Lond Ser B-Biol Sci. 1993;340(1292):167–177. doi: 10.1098/rstb.1993.0055. - DOI
-
- Lofstedt C, Vickers NJ, Baker TC. Courtship, Pheromone Titer and Determination of the Male Mating Success in the Oriental Fruit Moth, Grapholita-Molesta (Lepidoptera, Tortricidae) Entomologia Generalis. 1990;15(2):121–125.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

