A unique, thermostable dimer linkage structure of RD114 retroviral RNA
- PMID: 20075164
- PMCID: PMC2822922
- DOI: 10.1261/rna.1495110
A unique, thermostable dimer linkage structure of RD114 retroviral RNA
Abstract
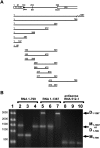
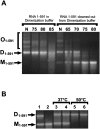
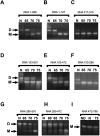
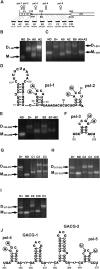
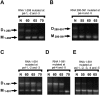
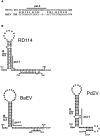
Retroviruses package their genome as RNA dimers linked together primarily by base-pairing between palindromic stem-loop (psl) sequences at the 5' end of genomic RNA. Retroviral RNA dimers usually melt in the range of 55 degrees C-70 degrees C. However, RNA dimers from virions of the feline endogenous gammaretrovirus RD114 were reported to melt only at 87 degrees C. We here report that the high thermal stability of RD114 RNA dimers generated from in vitro synthesized RNA is an effect of multiple dimerization sites located in the 5' region from the R region to sequences downstream from the splice donor (SD) site. By antisense oligonucleotide probing we were able to map at least five dimerization sites. Computational prediction revealed a possibility to form stems with autocomplementary loops for all of the mapped dimerization sites. Three of them were located upstream of the SD site. Mutant analysis supported a role of all five loop sequences in the formation and thermal stability of RNA dimers. Four of the five psls were also predicted in the RNA of two baboon endogenous retroviruses proposed to be ancestors of RD114. RNA fragments of the 5' R region or prolonged further downstream could be efficiently dimerized in vitro. However, this was not the case for the 3' R region linked to upstream U3 sequences, suggesting a specific mechanism of negative regulation of dimerization at the 3' end of the genome, possibly explained by a long double-stranded RNA region at the U3-R border. Altogether, these data point to determinants of the high thermostability of the dimer linkage structure of the RD114 genome and reveal differences from other retroviruses.
Figures

Similar articles
-
Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro.Proc Natl Acad Sci U S A. 1994 May 24;91(11):4945-9. doi: 10.1073/pnas.91.11.4945. Proc Natl Acad Sci U S A. 1994. PMID: 8197162 Free PMC article.
-
HIV-1 genome dimerization: kissing-loop hairpin dictates whether nucleotides downstream of the 5' splice junction contribute to loose and tight dimerization of human immunodeficiency virus RNA.Biochemistry. 1997 Aug 5;36(31):9501-8. doi: 10.1021/bi970862l. Biochemistry. 1997. PMID: 9235995
-
Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA.J Biol Chem. 1994 Nov 4;269(44):27486-93. J Biol Chem. 1994. PMID: 7961663
-
Bipartite signal for genomic RNA dimerization in Moloney murine leukemia virus.J Virol. 2002 Apr;76(7):3135-44. doi: 10.1128/jvi.76.7.3135-3144.2002. J Virol. 2002. PMID: 11884538 Free PMC article.
-
Dimerization of retroviral genomic RNAs: structural and functional implications.Biochimie. 1996;78(7):639-53. doi: 10.1016/s0300-9084(96)80010-1. Biochimie. 1996. PMID: 8955907 Review.
Cited by
-
Sequence comparison of three infectious molecular clones of RD-114 virus.Virus Genes. 2012 Oct;45(2):393-7. doi: 10.1007/s11262-012-0759-0. Epub 2012 May 26. Virus Genes. 2012. PMID: 22639102
-
Retroviral RNA Dimerization: From Structure to Functions.Front Microbiol. 2018 Mar 22;9:527. doi: 10.3389/fmicb.2018.00527. eCollection 2018. Front Microbiol. 2018. PMID: 29623074 Free PMC article. Review.
References
-
- Aagaard L, Rasmussen SV, Mikkelsen JG, Pedersen FS. Efficient replication of full-length murine leukemia viruses modified at the dimer initiation site regions. Virology. 2004;318:360–370. - PubMed
-
- Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J. Role of the trans-activation response element in dimerization of HIV-1 RNA. J Biol Chem. 2004;279:22243–22249. - PubMed
-
- Basyuk E, Boulon S, Skou Pedersen F, Bertrand E, Vestergaard Rasmussen S. The packaging signal of MLV is an integrated module that mediates intracellular transport of genomic RNAs. J Mol Biol. 2005;354:330–339. - PubMed
-
- Bender W, Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976;7:595–607. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
