Structure and activity of putative intronic miRNA promoters
- PMID: 20075166
- PMCID: PMC2822915
- DOI: 10.1261/rna.1731910
Structure and activity of putative intronic miRNA promoters
Abstract
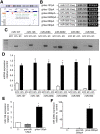
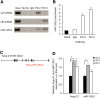
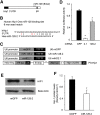
MicroRNAs (miRNAs) are RNA sequences of approximately 22 nucleotides that mediate post-transcriptional regulation of specific mRNAs. miRNA sequences are dispersed throughout the genome and are classified as intergenic (between genes) or intronic (embedded into a gene). Intergenic miRNAs are expressed by their own promoter, and until recently, it was supposed that intronic miRNAs are transcribed from their host gene. Here, we performed a genomic analysis of currently known intronic miRNA regions and observed that approximately 35% of intronic miRNAs have upstream regulatory elements consistent with promoter function. Among all intronic miRNAs, 30% have associated Pol II regulatory elements, including transcription start sites, CpG islands, expression sequence tags, and conserved transcription factor binding sites, while 5% contain RNA Pol III regulatory elements (A/B box sequences). We cloned intronic regions encompassing miRNAs and their upstream Pol II (miR-107, miR-126, miR-208b, miR-548f-2, miR-569, and miR-590) or Pol III (miR-566 and miR-128-2) sequences into a promoterless plasmid, and confirmed that miRNA expression occurs independent of host gene transcription. For miR-128-2, a miRNA overexpressed in acute lymphoblastic leukemia, ChIP analysis suggests dual regulation by both intronic (Pol III) and host gene (Pol II) promoters. These data support complex regulation of intronic miRNA expression, and have relevance to disregulation in disease settings.
Figures


References
-
- Agirre X, Novo FJ, Calasanz MJ, Larrayoz MJ, Lahortiga I, Valganon M, Garcia-Delgado M, Vizmanos JL. TP53 is frequently altered by methylation, mutation, and/or deletion in acute lymphoblastic leukaemia. Mol Carcinog. 2003;38:201–208. - PubMed
-
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
