Unique features of animal mitochondrial translation systems. The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases
- PMID: 20075606
- PMCID: PMC3417567
- DOI: 10.2183/pjab.86.11
Unique features of animal mitochondrial translation systems. The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases
Abstract
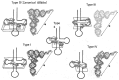
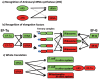
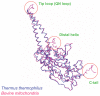
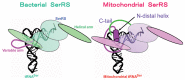
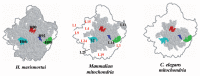
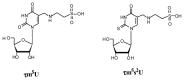
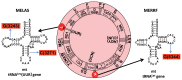
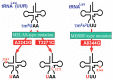
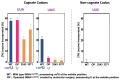
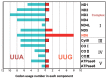
In animal mitochondria, several codons are non-universal and their meanings differ depending on the species. In addition, the tRNA structures that decipher codons are sometimes unusually truncated. These features seem to be related to the shortening of mitochondrial (mt) genomes, which occurred during the evolution of mitochondria. These organelles probably originated from the endosymbiosis of an aerobic eubacterium into an ancestral eukaryote. It is plausible that these events brought about the various characteristic features of animal mt translation systems, such as genetic code variations, unusually truncated tRNA and rRNA structures, unilateral tRNA recognition mechanisms by aminoacyl-tRNA synthetases, elongation factors and ribosomes, and compensation for RNA deficits by enlarged proteins. In this article, we discuss molecular mechanisms for these phenomena. Finally, we describe human mt diseases that are caused by modification defects in mt tRNAs.
Figures
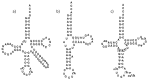



References
-
- Barrell B. G., Bankier A. T., Drouin J. (1979) A different genetic code in human mitochondria. Nature 282, 189–194 - PubMed
-
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H. L., Coulson A. R., Drouin J., et al. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 - PubMed
-
- Gesteland R. F., Ceck T. R., Atkins J. F. (eds.) (1999) The RNA World Second Edition – The nature of modern RNA suggests a prebiotic RNA world. Cold Spring Harbor Laboratory Press., New York, pp. 1–709

