Protacs for treatment of cancer
- PMID: 20075761
- PMCID: PMC2881331
- DOI: 10.1203/PDR.0b013e3181d35017
Protacs for treatment of cancer
Abstract
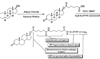
Protein degradation is the cell's mechanism of eliminating misfolded or unwanted proteins. The pathway by which proteins are degraded occurs through the ubiquitin-proteasome system. Ubiquitin is a small 9-kD (kDa) protein that is attached to proteins. A minimum of four ubiquitins are required for proteins to be recognized by the degradation machinery, known as the 26S proteasome. Defects in ubiquitination have been identified in a number of diseases, including cancer, neurodegenerative diseases, and metabolic disorders. We sought to exploit the delicate balance between protein synthesis and degradation to treat cancer by designing a chimeric molecule, known as Protac (Proteolysis Targeting Chimeric molecule). Protacs are heterobifunctional nanomolecules that are approximately 10 nm in size and can recruit proteins that cause cancer to the ubiquitin-proteasome machinery for degradation. In this review, we discuss the development of this novel technology for the treatment of cancer.
Figures




References
-
- Pray TR, Parlati F, Huang J, Wong BR, Payan DG, Bennett MK, Issakani SD, Molineaux S, Demo SD. Cell cycle regulatory E3 ubiquitin ligases as anticancer targets. Drug Resist Updat. 2002;5:249–258. - PubMed
-
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. - PubMed
-
- Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. - PubMed
-
- Sakamoto KM. Ubiquitin-dependent proteolysis: its role in human diseases and the design of therapeutic strategies. Mol Genet Metab. 2002;77:44–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

