Coordinated regulation of p53 apoptotic targets BAX and PUMA by SMAR1 through an identical MAR element
- PMID: 20075864
- PMCID: PMC2829167
- DOI: 10.1038/emboj.2009.395
Coordinated regulation of p53 apoptotic targets BAX and PUMA by SMAR1 through an identical MAR element
Abstract
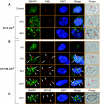
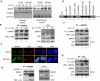
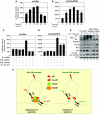
How tumour suppressor p53 bifurcates cell cycle arrest and apoptosis and executes these distinct pathways is not clearly understood. We show that BAX and PUMA promoters harbour an identical MAR element and are transcriptional targets of SMAR1. On mild DNA damage, SMAR1 selectively represses BAX and PUMA through binding to the MAR independently of inducing p53 deacetylation through HDAC1. This generates an anti-apoptotic response leading to cell cycle arrest. Importantly, knockdown of SMAR1 induces apoptosis, which is abrogated in the absence of p53. Conversely, apoptotic DNA damage results in increased size and number of promyelocytic leukaemia (PML) nuclear bodies with consequent sequestration of SMAR1. This facilitates p53 acetylation and restricts SMAR1 binding to BAX and PUMA MAR leading to apoptosis. Thus, our study establishes MAR as a damage responsive cis element and SMAR1-PML crosstalk as a switch that modulates the decision between cell cycle arrest and apoptosis in response to DNA damage.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
SMAR1 forms a ternary complex with p53-MDM2 and negatively regulates p53-mediated transcription.J Mol Biol. 2009 May 15;388(4):691-702. doi: 10.1016/j.jmb.2009.03.033. Epub 2009 Mar 19. J Mol Biol. 2009. PMID: 19303885
-
Chromatin remodelling protein SMAR1 inhibits p53 dependent transactivation by regulating acetyl transferase p300.Int J Biochem Cell Biol. 2012 Jan;44(1):46-52. doi: 10.1016/j.biocel.2011.10.020. Epub 2011 Nov 3. Int J Biochem Cell Biol. 2012. PMID: 22074660
-
Nuclear matrix protein SMAR1 represses HIV-1 LTR mediated transcription through chromatin remodeling.Virology. 2010 Apr 25;400(1):76-85. doi: 10.1016/j.virol.2010.01.017. Epub 2010 Feb 11. Virology. 2010. PMID: 20153010
-
Importance of proapoptotic protein PUMA in cell radioresistance.Folia Biol (Praha). 2014;60(2):53-6. doi: 10.14712/fb2014060020053. Folia Biol (Praha). 2014. PMID: 24785107 Review.
-
Biological functions of the ING family tumor suppressors.Cell Mol Life Sci. 2004 Oct;61(19-20):2597-613. doi: 10.1007/s00018-004-4199-4. Cell Mol Life Sci. 2004. PMID: 15526165 Free PMC article. Review.
Cited by
-
Curcumin enhances the efficacy of chemotherapy by tailoring p65NFκB-p300 cross-talk in favor of p53-p300 in breast cancer.J Biol Chem. 2011 Dec 9;286(49):42232-42247. doi: 10.1074/jbc.M111.262295. Epub 2011 Oct 19. J Biol Chem. 2011. PMID: 22013068 Free PMC article.
-
Nuclear matrix protein SMAR1 control regulatory T-cell fate during inflammatory bowel disease (IBD).Mucosal Immunol. 2015 Nov;8(6):1184-200. doi: 10.1038/mi.2015.42. Mucosal Immunol. 2015. PMID: 25993445 Free PMC article.
-
Connecting chromatin modifying factors to DNA damage response.Int J Mol Sci. 2013 Jan 24;14(2):2355-69. doi: 10.3390/ijms14022355. Int J Mol Sci. 2013. PMID: 23348929 Free PMC article.
-
GADD45β mediates p53 protein degradation via Src/PP2A/MDM2 pathway upon arsenite treatment.Cell Death Dis. 2013 May 16;4(5):e637. doi: 10.1038/cddis.2013.162. Cell Death Dis. 2013. PMID: 23681232 Free PMC article.
-
Tumor suppressor SMAR1 regulates PKM alternative splicing by HDAC6-mediated deacetylation of PTBP1.Cancer Metab. 2021 Apr 16;9(1):16. doi: 10.1186/s40170-021-00252-x. Cancer Metab. 2021. PMID: 33863392 Free PMC article.
References
-
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YM, Steen H, Mann M, Lamond AI (2002) Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 - PubMed
-
- Aylon Y, Oren M (2007) Living with p53, dying of p53. Cell 130: 597–600 - PubMed
-
- Bernardi R, Pandolfi PP (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8: 1006–1016 - PubMed
-
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP (2004) PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol 6: 665–672 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

