HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy
- PMID: 20075865
- PMCID: PMC2837169
- DOI: 10.1038/emboj.2009.405
HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy
Abstract
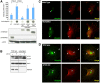
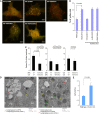
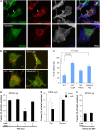
Autophagy is primarily considered a non-selective degradation process induced by starvation. Nutrient-independent basal autophagy, in contrast, imposes intracellular QC by selective disposal of aberrant protein aggregates and damaged organelles, a process critical for suppressing neurodegenerative diseases. The molecular mechanism that distinguishes these two fundamental autophagic responses, however, remains mysterious. Here, we identify the ubiquitin-binding deacetylase, histone deacetylase-6 (HDAC6), as a central component of basal autophagy that targets protein aggregates and damaged mitochondria. Surprisingly, HDAC6 is not required for autophagy activation; rather, it controls the fusion of autophagosomes to lysosomes. HDAC6 promotes autophagy by recruiting a cortactin-dependent, actin-remodelling machinery, which in turn assembles an F-actin network that stimulates autophagosome-lysosome fusion and substrate degradation. Indeed, HDAC6 deficiency leads to autophagosome maturation failure, protein aggregate build-up, and neurodegeneration. Remarkably, HDAC6 and F-actin assembly are completely dispensable for starvation-induced autophagy, uncovering the fundamental difference of these autophagic modes. Our study identifies HDAC6 and the actin cytoskeleton as critical components that define QC autophagy and uncovers a novel regulation of autophagy at the level of autophagosome-lysosome fusion.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
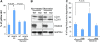
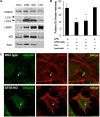
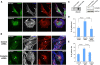

References
-
- Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z (2006) Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem 281: 36303–36316 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

