Regulation of antigen-receptor gene assembly in hagfish
- PMID: 20075989
- PMCID: PMC2828746
- DOI: 10.1038/embor.2009.274
Regulation of antigen-receptor gene assembly in hagfish
Abstract
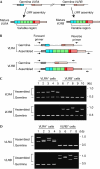
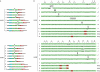
Variable lymphocyte receptors (VLRs) are antigen receptors in the jawless vertebrates lamprey and hagfish. VLR genes are classified into VLRA and VLRB, and lymphocytes expressing VLRA are T-cell-like, whereas those expressing VLRB are B-cell-like in the sea lamprey. Diverse VLR genes are assembled somatically in lymphocytes; however, how the assembly is regulated is still largely unknown. Here, we analyse VLR gene assembly at the single-cell level in the inshore hagfish (Eptatretus burgeri). Each lymphocyte assembles and transcribes only one type of VLR gene, either VLRA or VLRB. In general, monoallelic assembly of VLR was observed, but diallelic assembly was found in some cases--in many of which, one allele was functional and the other was defective. In fact, all VLR-assembled lymphocytes contained at least one functional VLR gene. Together, these results indicate a feedback inhibition of VLR assembly and selection of VLR-positive lymphocytes.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z (2005) Diversity and function of adaptive immune receptors in a jawless vertebrate. Science 310: 1970–1973 - PubMed
-
- Alder MN, Herrin BR, Sadlonova A, Stockard CR, Grizzle WE, Gartland LA, Gartland GL, Boydston JA, Turnbough CL Jr, Cooper MD (2008) Antibody responses of variable lymphocyte receptors in the lamprey. Nat Immunol 9: 319–327 - PubMed
-
- Bajoghli B et al. (2009) Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell 138: 186–197 - PubMed
-
- Cooper MD, Alder MN (2006) The evolution of adaptive immune systems. Cell 124: 815–822 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

