Genomics, evolution, and crystal structure of a new family of bacterial spore kinases
- PMID: 20077512
- PMCID: PMC2860764
- DOI: 10.1002/prot.22663
Genomics, evolution, and crystal structure of a new family of bacterial spore kinases
Abstract
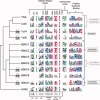
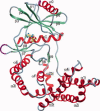
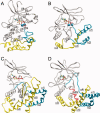
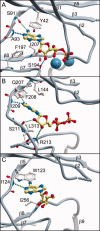
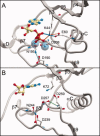
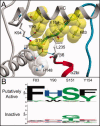
Bacterial spore formation is a complex process of fundamental relevance to biology and human disease. The spore coat structure is complex and poorly understood, and the roles of many of the protein components remain unclear. We describe a new family of spore coat proteins, the bacterial spore kinases (BSKs), and the first crystal structure of a BSK, YtaA (CotI) from Bacillus subtilis. BSKs are widely distributed in spore-forming Bacillus and Clostridium species, and have a dynamic evolutionary history. Sequence and structure analyses indicate that the BSKs are CAKs, a prevalent group of small molecule kinases in bacteria that is distantly related to the eukaryotic protein kinases. YtaA has substantial structural similarity to CAKs, but also displays distinctive features that broaden our understanding of the CAK group. Evolutionary constraint analysis of the protein surfaces indicates that members of the BSK family have distinct clade-conserved patterns in the substrate binding region, and probably bind and phosphorylate distinct targets. Several classes of BSKs have apparently independently lost catalytic activity to become pseudokinases, indicating that the family also has a major noncatalytic function.
2009 Wiley-Liss, Inc.
Figures


Similar articles
-
Diversity and evolutionary dynamics of spore-coat proteins in spore-forming species of Bacillales.Microb Genom. 2020 Nov;6(11):mgen000451. doi: 10.1099/mgen.0.000451. Epub 2020 Oct 14. Microb Genom. 2020. PMID: 33052805 Free PMC article.
-
Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes.Environ Microbiol. 2012 Nov;14(11):2870-90. doi: 10.1111/j.1462-2920.2012.02841.x. Epub 2012 Aug 13. Environ Microbiol. 2012. PMID: 22882546 Free PMC article.
-
Phosphorylation of spore coat proteins by a family of atypical protein kinases.Proc Natl Acad Sci U S A. 2016 Jun 21;113(25):E3482-91. doi: 10.1073/pnas.1605917113. Epub 2016 May 16. Proc Natl Acad Sci U S A. 2016. PMID: 27185916 Free PMC article.
-
Nucleotide-binding mechanisms in pseudokinases.Biosci Rep. 2015 Nov 20;36(1):e00282. doi: 10.1042/BSR20150226. Biosci Rep. 2015. PMID: 26589967 Free PMC article. Review.
-
Bacillus subtilis spore coat.Microbiol Mol Biol Rev. 1999 Mar;63(1):1-20. doi: 10.1128/MMBR.63.1.1-20.1999. Microbiol Mol Biol Rev. 1999. PMID: 10066829 Free PMC article. Review.
Cited by
-
Genome-wide identification and comprehensive analyses of the kinomes in four pathogenic microsporidia species.PLoS One. 2014 Dec 30;9(12):e115890. doi: 10.1371/journal.pone.0115890. eCollection 2014. PLoS One. 2014. PMID: 25549259 Free PMC article.
-
Identification and classification of small molecule kinases: insights into substrate recognition and specificity.BMC Evol Biol. 2016 Jan 6;16:7. doi: 10.1186/s12862-015-0576-x. BMC Evol Biol. 2016. PMID: 26738562 Free PMC article.
-
Recent progress in Bacillus subtilis sporulation.FEMS Microbiol Rev. 2012 Jan;36(1):131-48. doi: 10.1111/j.1574-6976.2011.00310.x. Epub 2011 Oct 25. FEMS Microbiol Rev. 2012. PMID: 22091839 Free PMC article. Review.
-
Diversity and evolutionary dynamics of spore-coat proteins in spore-forming species of Bacillales.Microb Genom. 2020 Nov;6(11):mgen000451. doi: 10.1099/mgen.0.000451. Epub 2020 Oct 14. Microb Genom. 2020. PMID: 33052805 Free PMC article.
-
The Bacillus subtilis endospore: assembly and functions of the multilayered coat.Nat Rev Microbiol. 2013 Jan;11(1):33-44. doi: 10.1038/nrmicro2921. Epub 2012 Dec 3. Nat Rev Microbiol. 2013. PMID: 23202530 Free PMC article. Review.
References
-
- Henriques AO, Moran CP., Jr Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol. 2007;61:555–588. - PubMed
-
- Setlow P. Spore germination. Curr Opin Microbiol. 2003;6:550–556. - PubMed
-
- Driks A. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol. 2002;10:251–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

