In silico and biological survey of transcription-associated proteins implicated in the transcriptional machinery during the erythrocytic development of Plasmodium falciparum
- PMID: 20078850
- PMCID: PMC2821373
- DOI: 10.1186/1471-2164-11-34
In silico and biological survey of transcription-associated proteins implicated in the transcriptional machinery during the erythrocytic development of Plasmodium falciparum
Abstract
Background: Malaria is the most important parasitic disease in the world with approximately two million people dying every year, mostly due to Plasmodium falciparum infection. During its complex life cycle in the Anopheles vector and human host, the parasite requires the coordinated and modulated expression of diverse sets of genes involved in epigenetic, transcriptional and post-transcriptional regulation. However, despite the availability of the complete sequence of the Plasmodium falciparum genome, we are still quite ignorant about Plasmodium mechanisms of transcriptional gene regulation. This is due to the poor prediction of nuclear proteins, cognate DNA motifs and structures involved in transcription.
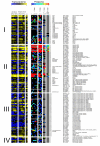
Results: A comprehensive directory of proteins reported to be potentially involved in Plasmodium transcriptional machinery was built from all in silico reports and databanks. The transcription-associated proteins were clustered in three main sets of factors: general transcription factors, chromatin-related proteins (structuring, remodelling and histone modifying enzymes), and specific transcription factors. Only a few of these factors have been molecularly analysed. Furthermore, from transcriptome and proteome data we modelled expression patterns of transcripts and corresponding proteins during the intra-erythrocytic cycle. Finally, an interactome of these proteins based either on in silico or on 2-yeast-hybrid experimental approaches is discussed.
Conclusion: This is the first attempt to build a comprehensive directory of potential transcription-associated proteins in Plasmodium. In addition, all complete transcriptome, proteome and interactome raw data were re-analysed, compared and discussed for a better comprehension of the complex biological processes of Plasmodium falciparum transcriptional regulation during the erythrocytic development.
Figures


References
-
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM. et al.Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

