The proteolytic system of lactic acid bacteria revisited: a genomic comparison
- PMID: 20078865
- PMCID: PMC2827410
- DOI: 10.1186/1471-2164-11-36
The proteolytic system of lactic acid bacteria revisited: a genomic comparison
Abstract
Background: Lactic acid bacteria (LAB) are a group of gram-positive, lactic acid producing Firmicutes. They have been extensively used in food fermentations, including the production of various dairy products. The proteolytic system of LAB converts proteins to peptides and then to amino acids, which is essential for bacterial growth and also contributes significantly to flavor compounds as end-products. Recent developments in high-throughput genome sequencing and comparative genomics hybridization arrays provide us with opportunities to explore the diversity of the proteolytic system in various LAB strains.
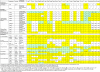
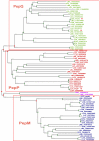
Results: We performed a genome-wide comparative genomics analysis of proteolytic system components, including cell-wall bound proteinase, peptide transporters and peptidases, in 22 sequenced LAB strains. The peptidase families PepP/PepQ/PepM, PepD and PepI/PepR/PepL are described as examples of our in silico approach to refine the distinction of subfamilies with different enzymatic activities. Comparison of protein 3D structures of proline peptidases PepI/PepR/PepL and esterase A allowed identification of a conserved core structure, which was then used to improve phylogenetic analysis and functional annotation within this protein superfamily.The diversity of proteolytic system components in 39 Lactococcus lactis strains was explored using pangenome comparative genome hybridization analysis. Variations were observed in the proteinase PrtP and its maturation protein PrtM, in one of the Opp transport systems and in several peptidases between strains from different Lactococcus subspecies or from different origin.
Conclusions: The improved functional annotation of the proteolytic system components provides an excellent framework for future experimental validations of predicted enzymatic activities. The genome sequence data can be coupled to other "omics" data e.g. transcriptomics and metabolomics for prediction of proteolytic and flavor-forming potential of LAB strains. Such an integrated approach can be used to tune the strain selection process in food fermentations.
Figures



References
-
- Kim YK, Yaguchi M, Rose D. Isolation and Amino Acid Composition of Para-Kappa-Casein. J Dairy Sci. 1969;52:316–320.
-
- Stewart AF, Bonsing J, Beattie CW, Shah F, Willis IM, Mackinlay AG. Complete nucleotide sequences of bovine alpha S2- and beta-casein cDNAs: comparisons with related sequences in other species. Mol Biol Evol. 1987;4:231–241. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

