Estrogen receptor-dependent regulation of CYP2B6 in human breast cancer cells
- PMID: 20079471
- PMCID: PMC5153331
- DOI: 10.1016/j.bbagrm.2010.01.005
Estrogen receptor-dependent regulation of CYP2B6 in human breast cancer cells
Abstract
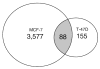
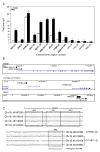
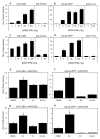
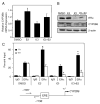
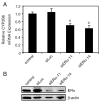
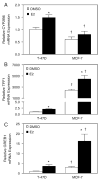
Estrogen receptor alpha (ERalpha) mediates the biological actions of estrogens and also contributes to the development and progression of breast cancer. To gain a more comprehensive understanding of ERalpha-mediated transcription, we used chromatin immunoprecipitation and promoter focused microarrays (ChIP-chip) to identify ERalpha binding sites in T-47D human breast cancer cells. Transcription factor binding site analysis revealed that the estrogen response element (ERE) was significantly over-represented and was found in 50% of the 243 ERalpha-bound regions identified. Interestingly, multiple ERalpha-bound regions were detected in the upstream regulatory sequences of the CYP2B gene cluster. Because ERalpha has been reported to regulate the expression of other cytochrome P450 enzymes and CYP2B6 is highly expressed in ERalpha-positive breast tumors, we focused on characterizing the ERalpha-dependent regulation of CYP2B6. Reporter gene assays revealed that ERalpha and ERbeta increased CYP2B6-regulated gene expression through a functional ERE located at -1669 to -1657 in the upstream regulatory region of CYP2B6. E2 increased ERalpha and nuclear receptor coactivator 3 (NCoA3) recruitment to the 5'-flanking region of CYP2B6, and increased CYP2B6 mRNA levels in T-47D but not in MCF-7 human breast cancer cells. RNAi-mediated knockdown of ERalpha in the T-47D cells resulted in a significant decrease in CYP2B6 mRNA levels. Taken together, our study provides evidence for cell-type specific transcriptional regulation of the CYP2B6 gene by ERs.
Figures


Similar articles
-
Phytoestrogens regulate mRNA and protein levels of guanine nucleotide-binding protein, beta-1 subunit (GNB1) in MCF-7 cells.J Cell Physiol. 2009 Jun;219(3):584-94. doi: 10.1002/jcp.21699. J Cell Physiol. 2009. PMID: 19170076 Free PMC article.
-
FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance.Oncogene. 2010 May 20;29(20):2983-95. doi: 10.1038/onc.2010.47. Epub 2010 Mar 8. Oncogene. 2010. PMID: 20208560 Free PMC article.
-
Estrogen receptor subtype- and promoter-specific modulation of aryl hydrocarbon receptor-dependent transcription.Mol Cancer Res. 2009 Jun;7(6):977-86. doi: 10.1158/1541-7786.MCR-08-0396. Epub 2009 May 26. Mol Cancer Res. 2009. PMID: 19470599
-
Regulation of specific target genes and biological responses by estrogen receptor subtype agonists.Curr Opin Pharmacol. 2010 Dec;10(6):629-36. doi: 10.1016/j.coph.2010.09.009. Epub 2010 Oct 14. Curr Opin Pharmacol. 2010. PMID: 20951642 Free PMC article. Review.
-
The role of SRC-3 in human breast cancer.Nat Rev Clin Oncol. 2010 Feb;7(2):83-9. doi: 10.1038/nrclinonc.2009.219. Epub 2009 Dec 22. Nat Rev Clin Oncol. 2010. PMID: 20027190 Review.
Cited by
-
Tumor-specific expression of microRNA-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and -independent pathways.Mol Ther. 2011 Aug;19(8):1521-8. doi: 10.1038/mt.2011.64. Epub 2011 May 24. Mol Ther. 2011. PMID: 21610700 Free PMC article.
-
Triclocarban mediates induction of xenobiotic metabolism through activation of the constitutive androstane receptor and the estrogen receptor alpha.PLoS One. 2012;7(6):e37705. doi: 10.1371/journal.pone.0037705. Epub 2012 Jun 15. PLoS One. 2012. PMID: 22761658 Free PMC article.
-
PARP7 and Mono-ADP-Ribosylation Negatively Regulate Estrogen Receptor α Signaling in Human Breast Cancer Cells.Cells. 2021 Mar 11;10(3):623. doi: 10.3390/cells10030623. Cells. 2021. PMID: 33799807 Free PMC article.
-
Bioactivation of the cancer chemopreventive agent tamoxifen to quinone methides by cytochrome P4502B6 and identification of the modified residue on the apoprotein.Drug Metab Dispos. 2012 Dec;40(12):2280-8. doi: 10.1124/dmd.112.047266. Epub 2012 Aug 31. Drug Metab Dispos. 2012. PMID: 22942317 Free PMC article.
-
The importance of both CYP2C19 and CYP2B6 germline variations in cyclophosphamide pharmacokinetics and clinical outcomes.Br J Clin Pharmacol. 2019 Sep;85(9):1925-1934. doi: 10.1111/bcp.14031. Epub 2019 Jul 22. Br J Clin Pharmacol. 2019. PMID: 31218720 Free PMC article. Review.
References
-
- Nilsson S, Gustafsson JA. Biological role of estrogen and estrogen receptors. Crit Rev Biochem Mol Biol. 2002;37:1–28. - PubMed
-
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. - PubMed
-
- Sanchez R, Nguyen D, Rocha W, White JH, Mader S. Diversity in the mechanisms of gene regulation by estrogen receptors. BioEssays. 2002;24:244–254. - PubMed
-
- Dutta U, Pant K. Aromatase inhibitors: past, present and future in breast cancer therapy. Med Oncol. 2008;25:113–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

