Development of quantitative and high-throughput assays of polyomavirus and papillomavirus DNA replication
- PMID: 20079917
- PMCID: PMC3154085
- DOI: 10.1016/j.virol.2009.12.026
Development of quantitative and high-throughput assays of polyomavirus and papillomavirus DNA replication
Abstract
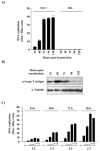
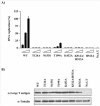
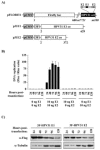
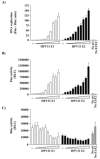
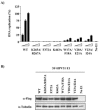
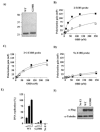
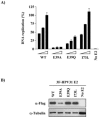
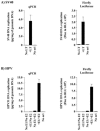
Polyoma- and papillomaviruses genome replication is initiated by the binding of large T antigen (LT) and of E1 and E2, respectively, at the viral origin (ori). Replication of an ori-containing plasmid occurs in cells transiently expressing these viral proteins and is typically quantified by Southern blotting or PCR. To facilitate the study of SV40 and HPV31 DNA replication, we developed cellular assays in which transient replication of the ori-plasmid is quantified using a firefly luciferase gene located in cis to the ori. Under optimized conditions, replication of the SV40 and HPV31 ori-plasmids resulted in a 50- and 150-fold increase in firefly luciferase levels, respectively. These results were validated using replication-defective mutants of LT, E1 and E2 and with inhibitors of DNA replication and cell-cycle progression. These quantitative and high-throughput assays should greatly facilitate the study of SV40 and HPV31 DNA replication and the identification of small-molecule inhibitors of this process.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
A quantitative and high-throughput assay of human papillomavirus DNA replication.Methods Mol Biol. 2015;1249:305-16. doi: 10.1007/978-1-4939-2013-6_23. Methods Mol Biol. 2015. PMID: 25348316
-
A high capacity assay for inhibitors of human papillomavirus DNA replication.Biotechnology (N Y). 1995 Nov;13(11):1210-4. doi: 10.1038/nbt1195-1210. Biotechnology (N Y). 1995. PMID: 9636294
-
[In vitro replication of SV40 and polyoma virus DNA].Tanpakushitsu Kakusan Koso. 1988 Sep;33(11):1766-73. Tanpakushitsu Kakusan Koso. 1988. PMID: 2853886 Japanese. No abstract available.
-
Role of mouse polyomavirus late region in the control of viral DNA replication: a review.Biochimie. 1995;77(10):780-6. doi: 10.1016/0300-9084(96)88196-x. Biochimie. 1995. PMID: 8824775 Review.
-
Cell alterations induced by the large T-antigens of SV40 and polyoma virus.Semin Cancer Biol. 1990 Dec;1(6):407-14. Semin Cancer Biol. 1990. PMID: 1966492 Review.
Cited by
-
Genetic variation of human papillomavirus type 16 in individual clinical specimens revealed by deep sequencing.PLoS One. 2013 Nov 13;8(11):e80583. doi: 10.1371/journal.pone.0080583. eCollection 2013. PLoS One. 2013. PMID: 24236186 Free PMC article.
-
Structure-based analysis of the interaction between the simian virus 40 T-antigen origin binding domain and single-stranded DNA.J Virol. 2011 Jan;85(2):818-27. doi: 10.1128/JVI.01738-10. Epub 2010 Oct 27. J Virol. 2011. PMID: 20980496 Free PMC article.
-
In Silico Identification of Potential Antagonists Targeting the HPV16 E2-E1 Interaction: A Step Toward Novel Therapeutics for Cervical Cancer.Curr Issues Mol Biol. 2025 Apr 18;47(4):288. doi: 10.3390/cimb47040288. Curr Issues Mol Biol. 2025. PMID: 40699687 Free PMC article.
-
Levels of the E2 interacting protein TopBP1 modulate papillomavirus maintenance stage replication.Virology. 2015 Apr;478:129-35. doi: 10.1016/j.virol.2015.01.011. Epub 2015 Feb 7. Virology. 2015. PMID: 25666521 Free PMC article.
-
The Replicative Consequences of Papillomavirus E2 Protein Binding to the Origin Replication Factor ORC2.PLoS Pathog. 2016 Oct 4;12(10):e1005934. doi: 10.1371/journal.ppat.1005934. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27701460 Free PMC article.
References
-
- Androphy EJ, Lowy DR, Schiller JT. Bovine papillomavirus E2 trans-activating gene product binds to specific sites in papillomavirus DNA. Nature. 1987;325(6099):70–3. - PubMed
-
- Auster AS, Joshua-Tor L. The DNA-binding domain of human papillomavirus type 18 E1. Crystal structure, dimerization, and DNA binding. J.Biol.Chem. 2004;279(5):3733–3742. - PubMed
-
- Bedin M, Gaben AM, Saucier C, Mester J. Geldanamycin, an inhibitor of the chaperone activity of HSP90, induces MAPK-independent cell cycle arrest. Int J Cancer. 2004;109(5):643–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

