Construction of an implicit membrane environment for the lattice Monte Carlo simulation of transmembrane protein
- PMID: 20079964
- PMCID: PMC7117040
- DOI: 10.1016/j.bpc.2009.12.008
Construction of an implicit membrane environment for the lattice Monte Carlo simulation of transmembrane protein
Abstract
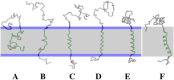
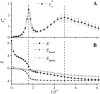
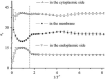
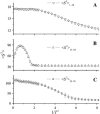
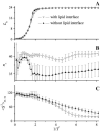
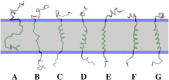
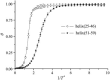
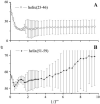
Due to the complexity of biological membrane, computer simulation of transmembrane protein's folding is challenging. In this paper, an implicit biological membrane environment has been constructed in lattice space, in which the lipid chains and water molecules were represented by the unoccupied lattice sites. The biological membrane was characterized with three features: stronger hydrogen bonding interaction, membrane lateral pressure, and lipophobicity index for the amino acid residues. In addition to the hydrocarbon core spanning region and the water solution, the lipid interface has also been represented in this implicit membrane environment, which was proved to be effective for the transmembrane protein's folding. The associated Monte Carlo simulations have been performed for SARS-CoV E protein and M2 protein segment (residues 18-60) of influenza A virus. It was found that the coil-helix transition of the transmembrane segment occurred earlier than the coil-globule transition of the two terminal domains. The folding process and final orientation of the amphipathic helical block in water solution are obviously influenced by its corresponding hydrophobicity/lipophobicity. Therefore, this implicit membrane environment, though in lattice space, can make an elaborate balance between different driving forces for the membrane protein's folding, thus offering a potential means for the simulation of transmembrane protein oligomers in feasible time.
Copyright 2009 Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Monte Carlo folding of trans-membrane helical peptides in an implicit generalized Born membrane.Proteins. 2007 Nov 1;69(2):297-308. doi: 10.1002/prot.21519. Proteins. 2007. PMID: 17600830
-
Computer simulations of membrane protein folding: structure and dynamics.Biophys J. 2003 Mar;84(3):1902-8. doi: 10.1016/S0006-3495(03)74998-4. Biophys J. 2003. PMID: 12609892 Free PMC article.
-
Spontaneous insertion of polypeptide chains into membranes: a Monte Carlo model.Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9391-5. doi: 10.1073/pnas.89.20.9391. Proc Natl Acad Sci U S A. 1992. PMID: 1409646 Free PMC article.
-
Marginally hydrophobic transmembrane α-helices shaping membrane protein folding.Protein Sci. 2015 Jul;24(7):1057-74. doi: 10.1002/pro.2698. Epub 2015 May 30. Protein Sci. 2015. PMID: 25970811 Free PMC article. Review.
-
Helix-helix interactions in membrane domains of bitopic proteins: Specificity and role of lipid environment.Biochim Biophys Acta Biomembr. 2017 Apr;1859(4):561-576. doi: 10.1016/j.bbamem.2016.10.024. Epub 2016 Nov 22. Biochim Biophys Acta Biomembr. 2017. PMID: 27884807 Review.
Cited by
-
Mechanism of function of viral channel proteins and implications for drug development.Int Rev Cell Mol Biol. 2012;294:259-321. doi: 10.1016/B978-0-12-394305-7.00006-9. Int Rev Cell Mol Biol. 2012. PMID: 22364876 Free PMC article. Review.
References
-
- White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomolec. Struct. 1999;28:319–365. - PubMed
-
- Bowie J.U. Solving the membrane protein folding problem. Nature. 2005;438:581–589. - PubMed
-
- Elofsson A., von Heijne G. Membrane protein structure: prediction versus reality. Annu. Rev. Biochem. 2007;76:125–140. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

