Metal templated design of protein interfaces
- PMID: 20080561
- PMCID: PMC2836610
- DOI: 10.1073/pnas.0906852107
Metal templated design of protein interfaces
Abstract
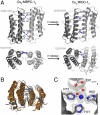
Metal coordination is a key structural and functional component of a large fraction of proteins. Given this dual role we considered the possibility that metal coordination may have played a templating role in the early evolution of protein folds and complexes. We describe here a rational design approach, Metal Templated Interface Redesign (MeTIR), that mimics the time course of a hypothetical evolutionary pathway for the formation of stable protein assemblies through an initial metal coordination event. Using a folded monomeric protein, cytochrome cb(562), as a building block we show that its non-self-associating surface can be made self-associating through a minimal number of mutations that enable Zn coordination. The protein interfaces in the resulting Zn-directed, D(2)-symmetrical tetramer are subsequently redesigned, yielding unique protein architectures that self-assemble in the presence or absence of metals. Aside from its evolutionary implications, MeTIR provides a route to engineer de novo protein interfaces and metal coordination environments that can be tuned through the extensive noncovalent bonding interactions in these interfaces.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
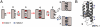
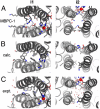

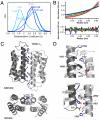
Comment in
-
Metal ions as matchmakers for proteins.Proc Natl Acad Sci U S A. 2010 Feb 2;107(5):1811-2. doi: 10.1073/pnas.0914008107. Proc Natl Acad Sci U S A. 2010. PMID: 20133828 Free PMC article. No abstract available.
Similar articles
-
Metal-Directed Design of Supramolecular Protein Assemblies.Methods Enzymol. 2016;580:223-50. doi: 10.1016/bs.mie.2016.05.009. Epub 2016 Jun 24. Methods Enzymol. 2016. PMID: 27586336 Free PMC article.
-
Metal-directed protein self-assembly.Acc Chem Res. 2010 May 18;43(5):661-72. doi: 10.1021/ar900273t. Acc Chem Res. 2010. PMID: 20192262 Free PMC article. Review.
-
Design and Construction of Functional Supramolecular Metalloprotein Assemblies.Acc Chem Res. 2019 Feb 19;52(2):345-355. doi: 10.1021/acs.accounts.8b00617. Epub 2019 Jan 30. Acc Chem Res. 2019. PMID: 30698941
-
De Novo Design of an Allosteric Metalloprotein Assembly with Strained Disulfide Bonds.J Am Chem Soc. 2016 Oct 12;138(40):13163-13166. doi: 10.1021/jacs.6b08458. Epub 2016 Sep 27. J Am Chem Soc. 2016. PMID: 27649076 Free PMC article.
-
Metals at the Helm: Revolutionizing Protein Assembly and Applications.Macromol Biosci. 2024 Nov;24(11):e2400126. doi: 10.1002/mabi.202400126. Epub 2024 Sep 6. Macromol Biosci. 2024. PMID: 39239781 Review.
Cited by
-
Recognition of functional roles of free radicals.Curr Neuropharmacol. 2012 Dec;10(4):287-8. doi: 10.2174/157015912804499474. Curr Neuropharmacol. 2012. PMID: 23730252 Free PMC article. No abstract available.
-
Confirmation of intersubunit connectivity and topology of designed protein complexes by native MS.Proc Natl Acad Sci U S A. 2018 Feb 6;115(6):1268-1273. doi: 10.1073/pnas.1713646115. Epub 2018 Jan 19. Proc Natl Acad Sci U S A. 2018. PMID: 29351988 Free PMC article.
-
Using anchoring motifs for the computational design of protein-protein interactions.Biochem Soc Trans. 2013 Oct;41(5):1141-5. doi: 10.1042/BST20130108. Biochem Soc Trans. 2013. PMID: 24059499 Free PMC article. Review.
-
Quantifying structural relationships of metal-binding sites suggests origins of biological electron transfer.Sci Adv. 2022 Jan 14;8(2):eabj3984. doi: 10.1126/sciadv.abj3984. Epub 2022 Jan 14. Sci Adv. 2022. PMID: 35030025 Free PMC article.
-
Self-assembled two-dimensional protein arrays in bionanotechnology: from S-layers to designed lattices.Curr Opin Biotechnol. 2014 Aug;28:39-45. doi: 10.1016/j.copbio.2013.11.001. Epub 2013 Nov 25. Curr Opin Biotechnol. 2014. PMID: 24832073 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

