Posttranslational regulation impacts the fate of duplicated genes
- PMID: 20080574
- PMCID: PMC2840353
- DOI: 10.1073/pnas.0911603107
Posttranslational regulation impacts the fate of duplicated genes
Abstract
Gene and genome duplications create novel genetic material on which evolution can work and have therefore been recognized as a major source of innovation for many eukaryotic lineages. Following duplication, the most likely fate is gene loss; however, a considerable fraction of duplicated genes survive. Not all genes have the same probability of survival, but it is not fully understood what evolutionary forces determine the pattern of gene retention. Here, we use genome sequence data as well as large-scale phosphoproteomics data from the baker's yeast Saccharomyces cerevisiae, which underwent a whole-genome duplication approximately 100 mya, and show that the number of phosphorylation sites on the proteins they encode is a major determinant of gene retention. Protein phosphorylation motifs are short amino acid sequences that are usually embedded within unstructured and rapidly evolving protein regions. Reciprocal loss of those ancestral sites and the gain of new ones are major drivers in the retention of the two surviving duplicates and in their acquisition of distinct functions. This way, small changes in the sequences of unstructured regions in proteins can contribute to the rapid rewiring and adaptation of regulatory networks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
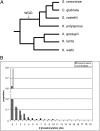
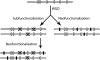

References
-
- Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. - PubMed
-
- Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970.
-
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. - PubMed
-
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

