HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres
- PMID: 20080577
- PMCID: PMC2824361
- DOI: 10.1073/pnas.0913709107
HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres
Abstract
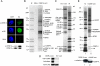
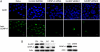
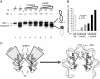
The human histone H3 variant, CENP-A, replaces the conventional histone H3 in centromeric chromatin and, together with centromere-specific DNA-binding factors, directs the assembly of the kinetochore. We purified the prenucelosomal e-CENP-A complex. We found that HJURP, a member of the complex, was required for cell cycle specific targeting of CENP-A to centromeres. HJURP facilitated efficient deposition of CENP-A/H4 tetramers to naked DNA in vitro. Bacterially expressed HJURP binds at a stoichiometric ratio to the CENP-A/H4 tetramer but not to the H3/H4 tetramer. The binding occurred through a conserved HJURP short N-terminal domain, termed CBD. The novel characteristic identified in vertebrates that we named TLTY box of CBD, was essential for formation of the HJURP-CENP-A/H4 complex. Our data identified HJURP as a vertebrate CENP-A chaperone and dissected its mode of interactions with CENP-A.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition.EMBO J. 2013 Jul 31;32(15):2113-24. doi: 10.1038/emboj.2013.142. Epub 2013 Jun 14. EMBO J. 2013. PMID: 23771058 Free PMC article.
-
Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP.Genes Dev. 2011 May 1;25(9):901-6. doi: 10.1101/gad.2045111. Epub 2011 Apr 8. Genes Dev. 2011. PMID: 21478274 Free PMC article.
-
HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly.Dev Cell. 2012 Apr 17;22(4):749-62. doi: 10.1016/j.devcel.2012.02.001. Epub 2012 Mar 8. Dev Cell. 2012. PMID: 22406139 Free PMC article.
-
Putting CENP-A in its place.Cell Mol Life Sci. 2013 Feb;70(3):387-406. doi: 10.1007/s00018-012-1048-8. Epub 2012 Jun 23. Cell Mol Life Sci. 2013. PMID: 22729156 Free PMC article. Review.
-
The histone variant CENP-A and centromere specification.Curr Opin Cell Biol. 2008 Feb;20(1):91-100. doi: 10.1016/j.ceb.2007.11.007. Epub 2008 Jan 15. Curr Opin Cell Biol. 2008. PMID: 18226513 Review.
Cited by
-
A pan-cancer analysis of the oncogenic role of Holliday junction recognition protein in human tumors.Open Med (Wars). 2022 Feb 16;17(1):317-328. doi: 10.1515/med-2022-0423. eCollection 2022. Open Med (Wars). 2022. PMID: 35274047 Free PMC article.
-
Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids.Proc Natl Acad Sci U S A. 2011 Aug 16;108(33):E498-505. doi: 10.1073/pnas.1103190108. Epub 2011 Jul 11. Proc Natl Acad Sci U S A. 2011. PMID: 21746892 Free PMC article.
-
Prognostic value of Holliday junction-recognizing protein and its correlation with immune infiltrates in lung adenocarcinoma.Oncol Lett. 2022 May 27;24(1):232. doi: 10.3892/ol.2022.13353. eCollection 2022 Jul. Oncol Lett. 2022. PMID: 35720487 Free PMC article.
-
Holliday Cross-Recognition Protein HJURP: Association With the Tumor Microenvironment in Hepatocellular Carcinoma and With Patient Prognosis.Pathol Oncol Res. 2022 Jun 17;28:1610506. doi: 10.3389/pore.2022.1610506. eCollection 2022. Pathol Oncol Res. 2022. PMID: 35783358 Free PMC article.
-
Mitotic regulator Mis18β interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP).J Biol Chem. 2014 Mar 21;289(12):8326-36. doi: 10.1074/jbc.M113.529958. Epub 2014 Feb 11. J Biol Chem. 2014. PMID: 24519934 Free PMC article.
References
-
- Mitelman F. Catalog of Chromosome Aberrations in Cancer. 5th Ed. New York: Wiley; 1994.
-
- Henikoff S, Dalal Y. Centromeric chromatin: What makes it unique? Curr Opin Genet Dev. 2005;15:177–184. - PubMed
-
- Cooper JL, Henikoff S. Adaptive evolution of the histone fold domain in centromeric histones. Mol Biol Evol. 2004;21:1712–1718. - PubMed
-
- Smith MM. Centromeres and variant histones: What, where, when, and why? Curr Opin Cell Biol. 2002;14:279–285. - PubMed
-
- Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

