Rate, molecular spectrum, and consequences of human mutation
- PMID: 20080596
- PMCID: PMC2824313
- DOI: 10.1073/pnas.0912629107
Rate, molecular spectrum, and consequences of human mutation
Abstract
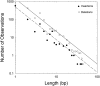
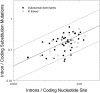
Although mutation provides the fuel for phenotypic evolution, it also imposes a substantial burden on fitness through the production of predominantly deleterious alleles, a matter of concern from a human-health perspective. Here, recently established databases on de novo mutations for monogenic disorders are used to estimate the rate and molecular spectrum of spontaneously arising mutations and to derive a number of inferences with respect to eukaryotic genome evolution. Although the human per-generation mutation rate is exceptionally high, on a per-cell division basis, the human germline mutation rate is lower than that recorded for any other species. Comparison with data from other species demonstrates a universal mutational bias toward A/T composition, and leads to the hypothesis that genome-wide nucleotide composition generally evolves to the point at which the power of selection in favor of G/C is approximately balanced by the power of random genetic drift, such that variation in equilibrium genome-wide nucleotide composition is largely defined by variation in mutation biases. Quantification of the hazards associated with introns reveals that mutations at key splice-site residues are a major source of human mortality. Finally, a consideration of the long-term consequences of current human behavior for deleterious-mutation accumulation leads to the conclusion that a substantial reduction in human fitness can be expected over the next few centuries in industrialized societies unless novel means of genetic intervention are developed.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

