Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands
- PMID: 20080605
- PMCID: PMC2824364
- DOI: 10.1073/pnas.0914163107
Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands
Abstract
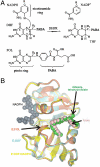
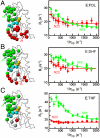
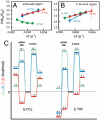
Enzyme catalysis can be described as progress over a multi-dimensional energy landscape where ensembles of interconverting conformational substates channel the enzyme through its catalytic cycle. We applied NMR relaxation dispersion to investigate the role of bound ligands in modulating the dynamics and energy landscape of Escherichia coli dihydrofolate reductase to obtain insights into the mechanism by which the enzyme efficiently samples functional conformations as it traverses its reaction pathway. Although the structural differences between the occluded substrate binary complexes and product ternary complexes are very small, there are substantial differences in protein dynamics. Backbone fluctuations on the micros-ms timescale in the cofactor binding cleft are similar for the substrate and product binary complexes, but fluctuations on this timescale in the active site loops are observed only for complexes with substrate or substrate analog and are not observed for the binary product complex. The dynamics in the substrate and product binary complexes are governed by quite different kinetic and thermodynamic parameters. Analogous dynamic differences in the E:THF:NADPH and E:THF:NADP(+) product ternary complexes are difficult to rationalize from ground-state structures. For both of these complexes, the nicotinamide ring resides outside the active site pocket in the ground state. However, they differ in the structure, energetics, and dynamics of accessible higher energy substates where the nicotinamide ring transiently occupies the active site. Overall, our results suggest that dynamics in dihydrofolate reductase are exquisitely "tuned" for every intermediate in the catalytic cycle; structural fluctuations efficiently channel the enzyme through functionally relevant conformational space.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
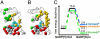
Similar articles
-
Cofactor-Mediated Conformational Dynamics Promote Product Release From Escherichia coli Dihydrofolate Reductase via an Allosteric Pathway.J Am Chem Soc. 2015 Jul 29;137(29):9459-68. doi: 10.1021/jacs.5b05707. Epub 2015 Jul 14. J Am Chem Soc. 2015. PMID: 26147643 Free PMC article.
-
The dynamic energy landscape of dihydrofolate reductase catalysis.Science. 2006 Sep 15;313(5793):1638-42. doi: 10.1126/science.1130258. Science. 2006. PMID: 16973882
-
Conformational changes in the active site loops of dihydrofolate reductase during the catalytic cycle.Biochemistry. 2004 Dec 28;43(51):16046-55. doi: 10.1021/bi048119y. Biochemistry. 2004. PMID: 15609999
-
Searching sequence space: two different approaches to dihydrofolate reductase catalysis.Chembiochem. 2005 Apr;6(4):590-600. doi: 10.1002/cbic.200400237. Chembiochem. 2005. PMID: 15812782 Review.
-
Conformational Sub-states and Populations in Enzyme Catalysis.Methods Enzymol. 2016;578:273-97. doi: 10.1016/bs.mie.2016.05.023. Epub 2016 Jul 9. Methods Enzymol. 2016. PMID: 27497171 Free PMC article. Review.
Cited by
-
Enzyme dynamics from NMR spectroscopy.Acc Chem Res. 2015 Feb 17;48(2):457-65. doi: 10.1021/ar500340a. Epub 2015 Jan 9. Acc Chem Res. 2015. PMID: 25574774 Free PMC article. Review.
-
Cofactor-Mediated Conformational Dynamics Promote Product Release From Escherichia coli Dihydrofolate Reductase via an Allosteric Pathway.J Am Chem Soc. 2015 Jul 29;137(29):9459-68. doi: 10.1021/jacs.5b05707. Epub 2015 Jul 14. J Am Chem Soc. 2015. PMID: 26147643 Free PMC article.
-
Evolutionarily conserved linkage between enzyme fold, flexibility, and catalysis.PLoS Biol. 2011 Nov;9(11):e1001193. doi: 10.1371/journal.pbio.1001193. Epub 2011 Nov 8. PLoS Biol. 2011. PMID: 22087074 Free PMC article.
-
Enzymatic Kinetic Isotope Effects from Path-Integral Free Energy Perturbation Theory.Methods Enzymol. 2016;577:359-88. doi: 10.1016/bs.mie.2016.05.057. Epub 2016 Jul 22. Methods Enzymol. 2016. PMID: 27498645 Free PMC article.
-
Redox-linked domain movements in the catalytic cycle of cytochrome p450 reductase.Structure. 2013 Sep 3;21(9):1581-9. doi: 10.1016/j.str.2013.06.022. Epub 2013 Aug 1. Structure. 2013. PMID: 23911089 Free PMC article.
References
-
- Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. - PubMed
-
- Bryngelson JD, Wolynes PG. Intermediates and barrier crossing in a random energy model (with applications to protein folding) J Phys Chem. 1989;93:6902–6915.
-
- Lazaridis T, Karplus M. "New view" of protein folding reconciled with the old through multiple unfolding simulations. Science. 1997;278:1928–1931. - PubMed
-
- Fernandez-Busquets X, de Groot NS, Fernandez D, Ventura S. Recent structural and computational insights into conformational diseases. Curr Med Chem. 2008;15:1336–1349. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

