Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3' UTR of turnip crinkle virus
- PMID: 20080629
- PMCID: PMC2803139
- DOI: 10.1073/pnas.0908140107
Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3' UTR of turnip crinkle virus
Abstract
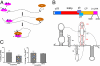
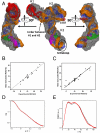
The 3(') untranslated region (3(') UTR) of turnip crinkle virus (TCV) genomic RNA contains a cap-independent translation element (CITE), which includes a ribosome-binding structural element (RBSE) that participates in recruitment of the large ribosomal subunit. In addition, a large symmetric loop in the RBSE plays a key role in coordinating the incompatible processes of viral translation and replication, which require enzyme progression in opposite directions on the viral template. To understand the structural basis for the large ribosomal subunit recruitment and the intricate interplay among different parts of the molecule, we determined the global structure of the 102-nt RBSE RNA using solution NMR and small-angle x-ray scattering. This RNA has many structural features that resemble those of a tRNA in solution. The hairpins H1 and H2, linked by a 7-nucleotide linker, form the upper part of RBSE and hairpin H3 is relatively independent from the rest of the structure and is accessible to interactions. This global structure provides insights into the three-dimensional layout for ribosome binding, which may serve as a structural basis for its involvement in recruitment of the large ribosomal subunit and the switch between viral translation and replication. The experimentally determined three-dimensional structure of a functional element in the 3(') UTR of an RNA from any organism has not been previously reported. The RBSE structure represents a prototype structure of a new class of RNA structural elements involved in viral translation/replication processes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

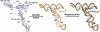
Similar articles
-
The 3' proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits.RNA. 2008 Nov;14(11):2379-93. doi: 10.1261/rna.1227808. Epub 2008 Sep 29. RNA. 2008. PMID: 18824512 Free PMC article.
-
Ribosome binding to a 5' translational enhancer is altered in the presence of the 3' untranslated region in cap-independent translation of turnip crinkle virus.J Virol. 2011 May;85(10):4638-53. doi: 10.1128/JVI.00005-11. Epub 2011 Mar 9. J Virol. 2011. PMID: 21389125 Free PMC article.
-
A ribosome-binding, 3' translational enhancer has a T-shaped structure and engages in a long-distance RNA-RNA interaction.J Virol. 2012 Sep;86(18):9828-42. doi: 10.1128/JVI.00677-12. Epub 2012 Jul 3. J Virol. 2012. PMID: 22761367 Free PMC article.
-
3'UTRs of carmoviruses.Virus Res. 2015 Aug 3;206:27-36. doi: 10.1016/j.virusres.2015.01.023. Epub 2015 Feb 4. Virus Res. 2015. PMID: 25662021 Review.
-
Functions of the 5'- and 3'-untranslated regions of tobamovirus RNA.Virus Res. 2015 Aug 3;206:82-9. doi: 10.1016/j.virusres.2015.01.028. Epub 2015 Feb 12. Virus Res. 2015. PMID: 25683511 Review.
Cited by
-
Predicting translational diffusion of evolutionary conserved RNA structures by the nucleotide number.Nucleic Acids Res. 2011 Feb;39(3):e17. doi: 10.1093/nar/gkq808. Epub 2010 Nov 10. Nucleic Acids Res. 2011. PMID: 21068070 Free PMC article.
-
Evidence of pervasive biologically functional secondary structures within the genomes of eukaryotic single-stranded DNA viruses.J Virol. 2014 Feb;88(4):1972-89. doi: 10.1128/JVI.03031-13. Epub 2013 Nov 27. J Virol. 2014. PMID: 24284329 Free PMC article.
-
Synthesis and applications of RNAs with position-selective labelling and mosaic composition.Nature. 2015 Jun 18;522(7556):368-72. doi: 10.1038/nature14352. Epub 2015 May 4. Nature. 2015. PMID: 25938715 Free PMC article.
-
Small-angle X-ray scattering-derived structure of the HIV-1 5' UTR reveals 3D tRNA mimicry.Proc Natl Acad Sci U S A. 2014 Mar 4;111(9):3395-400. doi: 10.1073/pnas.1319658111. Epub 2014 Feb 18. Proc Natl Acad Sci U S A. 2014. PMID: 24550473 Free PMC article.
-
Non-Canonical Translation Initiation Mechanisms Employed by Eukaryotic Viral mRNAs.Biochemistry (Mosc). 2021 Sep;86(9):1060-1094. doi: 10.1134/S0006297921090042. Biochemistry (Mosc). 2021. PMID: 34565312 Free PMC article. Review.
References
-
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 234:187–208. - PubMed
-
- Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. - PubMed
-
- Tarun SZ, Jr, Sachs AB. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. - PubMed
-
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

