Tracking footprints of artificial selection in the dog genome
- PMID: 20080661
- PMCID: PMC2824266
- DOI: 10.1073/pnas.0909918107
Tracking footprints of artificial selection in the dog genome
Abstract
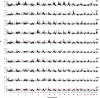
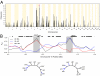
The size, shape, and behavior of the modern domesticated dog has been sculpted by artificial selection for at least 14,000 years. The genetic substrates of selective breeding, however, remain largely unknown. Here, we describe a genome-wide scan for selection in 275 dogs from 10 phenotypically diverse breeds that were genotyped for over 21,000 autosomal SNPs. We identified 155 genomic regions that possess strong signatures of recent selection and contain candidate genes for phenotypes that vary most conspicuously among breeds, including size, coat color and texture, behavior, skeletal morphology, and physiology. In addition, we demonstrate a significant association between HAS2 and skin wrinkling in the Shar-Pei, and provide evidence that regulatory evolution has played a prominent role in the phenotypic diversification of modern dog breeds. Our results provide a first-generation map of selection in the dog, illustrate how such maps can rapidly inform the genetic basis of canine phenotypic variation, and provide a framework for delineating the mechanistic basis of how artificial selection promotes rapid and pronounced phenotypic evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping.PLoS Genet. 2011 Oct;7(10):e1002316. doi: 10.1371/journal.pgen.1002316. Epub 2011 Oct 13. PLoS Genet. 2011. PMID: 22022279 Free PMC article.
-
A novel unstable duplication upstream of HAS2 predisposes to a breed-defining skin phenotype and a periodic fever syndrome in Chinese Shar-Pei dogs.PLoS Genet. 2011 Mar;7(3):e1001332. doi: 10.1371/journal.pgen.1001332. Epub 2011 Mar 17. PLoS Genet. 2011. PMID: 21437276 Free PMC article.
-
Linked genetic variants on chromosome 10 control ear morphology and body mass among dog breeds.BMC Genomics. 2015 Jun 23;16(1):474. doi: 10.1186/s12864-015-1702-2. BMC Genomics. 2015. PMID: 26100605 Free PMC article.
-
Insights into morphology and disease from the dog genome project.Annu Rev Cell Dev Biol. 2014;30:535-60. doi: 10.1146/annurev-cellbio-100913-012927. Epub 2014 Jul 9. Annu Rev Cell Dev Biol. 2014. PMID: 25062362 Free PMC article. Review.
-
Copy number variation in the domestic dog.Mamm Genome. 2012 Feb;23(1-2):144-63. doi: 10.1007/s00335-011-9369-8. Epub 2011 Dec 4. Mamm Genome. 2012. PMID: 22138850 Review.
Cited by
-
Origins of the domestic dog and the rich potential for gene mapping.Genet Res Int. 2011;2011:579308. doi: 10.4061/2011/579308. Epub 2011 Jan 17. Genet Res Int. 2011. PMID: 22567358 Free PMC article.
-
Whole Genome Resequencing Reveals Selection Signatures Associated With Important Traits in Ethiopian Indigenous Goat Populations.Front Genet. 2019 Nov 28;10:1190. doi: 10.3389/fgene.2019.01190. eCollection 2019. Front Genet. 2019. PMID: 31850061 Free PMC article.
-
Improved haplotype-based detection of ongoing selective sweeps towards an application in Arabidopsis thaliana.BMC Res Notes. 2011 Jul 5;4:232. doi: 10.1186/1756-0500-4-232. BMC Res Notes. 2011. PMID: 21729283 Free PMC article.
-
Impact of natural selection on global patterns of genetic variation, and association with clinical phenotypes, at genes involved in SARS-CoV-2 infection.medRxiv [Preprint]. 2021 Aug 7:2021.06.28.21259529. doi: 10.1101/2021.06.28.21259529. medRxiv. 2021. Update in: Proc Natl Acad Sci U S A. 2022 May 24;119(21):e2123000119. doi: 10.1073/pnas.2123000119. PMID: 34230933 Free PMC article. Updated. Preprint.
-
Demographic history, selection and functional diversity of the canine genome.Nat Rev Genet. 2017 Dec;18(12):705-720. doi: 10.1038/nrg.2017.67. Epub 2017 Sep 25. Nat Rev Genet. 2017. PMID: 28944780 Review.
References
-
- American Kennel Club . In: The Complete Dog Book. American Kennel Club Staff, editor. Foster City: Howell Book House; 1998.
-
- Sutter NB, Ostrander EA. Dog star rising: the canine genetic system. Nat Rev Genet. 2004;5:900–910. - PubMed
-
- Neff MW, Rine J. A fetching model organism. Cell. 2006;124:229–231. - PubMed
-
- Vilà C, et al. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. - PubMed
-
- Leonard JA, et al. Ancient DNA evidence for Old World origin of New World dogs. Science. 2002;298:1613–1616. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

