Evolution in health and medicine Sackler colloquium: Genomic disorders: a window into human gene and genome evolution
- PMID: 20080665
- PMCID: PMC2868291
- DOI: 10.1073/pnas.0906222107
Evolution in health and medicine Sackler colloquium: Genomic disorders: a window into human gene and genome evolution
Abstract
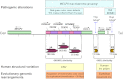
Gene duplications alter the genetic constitution of organisms and can be a driving force of molecular evolution in humans and the great apes. In this context, the study of genomic disorders has uncovered the essential role played by the genomic architecture, especially low copy repeats (LCRs) or segmental duplications (SDs). In fact, regardless of the mechanism, LCRs can mediate or stimulate rearrangements, inciting genomic instability and generating dynamic and unstable regions prone to rapid molecular evolution. In humans, copy-number variation (CNV) has been implicated in common traits such as neuropathy, hypertension, color blindness, infertility, and behavioral traits including autism and schizophrenia, as well as disease susceptibility to HIV, lupus nephritis, and psoriasis among many other clinical phenotypes. The same mechanisms implicated in the origin of genomic disorders may also play a role in the emergence of segmental duplications and the evolution of new genes by means of genomic and gene duplication and triplication, exon shuffling, exon accretion, and fusion/fission events.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Serial segmental duplications during primate evolution result in complex human genome architecture.Genome Res. 2004 Nov;14(11):2209-20. doi: 10.1101/gr.2746604. Genome Res. 2004. PMID: 15520286 Free PMC article.
-
Inverted low-copy repeats and genome instability--a genome-wide analysis.Hum Mutat. 2013 Jan;34(1):210-20. doi: 10.1002/humu.22217. Epub 2012 Oct 11. Hum Mutat. 2013. PMID: 22965494 Free PMC article.
-
Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs.Genome Res. 2002 May;12(5):729-38. doi: 10.1101/gr.82802. Genome Res. 2002. PMID: 11997339 Free PMC article.
-
Genomic disorders: genome architecture results in susceptibility to DNA rearrangements causing common human traits.Cold Spring Harb Symp Quant Biol. 2003;68:445-54. doi: 10.1101/sqb.2003.68.445. Cold Spring Harb Symp Quant Biol. 2003. PMID: 15338647 Review. No abstract available.
-
Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes.PLoS Genet. 2005 Dec;1(6):e49. doi: 10.1371/journal.pgen.0010049. PLoS Genet. 2005. PMID: 16444292 Free PMC article. Review.
Cited by
-
On the spot: very local chromosomal rearrangements.F1000 Biol Rep. 2012;4:22. doi: 10.3410/B4-22. Epub 2012 Nov 1. F1000 Biol Rep. 2012. PMID: 23189093 Free PMC article.
-
Genes that are Used Together are More Likely to be Fused Together in Evolution by Mutational Mechanisms: A Bioinformatic Test of the Used-Fused Hypothesis.Evol Biol. 2023;50(1):30-55. doi: 10.1007/s11692-022-09579-9. Epub 2022 Nov 30. Evol Biol. 2023. PMID: 36816837 Free PMC article.
-
Detection of genetic alterations in gastric cancer patients from Saudi Arabia using comparative genomic hybridization (CGH).PLoS One. 2018 Sep 13;13(9):e0202576. doi: 10.1371/journal.pone.0202576. eCollection 2018. PLoS One. 2018. PMID: 30212456 Free PMC article.
-
Structural variation-associated expression changes are paralleled by chromatin architecture modifications.PLoS One. 2013 Nov 12;8(11):e79973. doi: 10.1371/journal.pone.0079973. eCollection 2013. PLoS One. 2013. PMID: 24265791 Free PMC article.
-
Genomic Variation, Evolvability, and the Paradox of Mental Illness.Front Psychiatry. 2021 Jan 21;11:593233. doi: 10.3389/fpsyt.2020.593233. eCollection 2020. Front Psychiatry. 2021. PMID: 33551865 Free PMC article.
References
-
- Lupski JR. Genomic disorders: Structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

