Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion
- PMID: 20080673
- PMCID: PMC2836689
- DOI: 10.1073/pnas.0913396107
Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion
Abstract
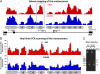
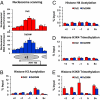
Plasmodium falciparum can invade erythrocytes by redundant receptors, some of which have variable expression. A P. falciparum clone Dd2 requiring erythrocyte sialic acid for invasion can be switched to a sialic acid-independent progeny clone Dd2NM by growing the Dd2 clone with neuraminidase-treated erythrocytes. The RH4 gene is transcriptionally up-regulated in Dd2NM compared to Dd2, despite the absence of DNA changes in and around the gene. We determined the epigenetic modifications around the transcription start site (TSS) at the time of expression of RH4 in Dd2NM (44 h) and at an earlier time when RH4 is not expressed (24 h). At 44 h, the occupancy of the +1 nucleosome site downstream of the TSS of the active RH4 gene in Dd2NM was markedly reduced compared to Dd2; no difference was observed at 24 h. At 44 h, histone modifications associated with up-regulation were positively correlated to the active RH4 gene of Dd2NM compared to Dd2; no differences were observed at 24 h. Histone H3K9 trimethylation (a marker for silencing) was higher in Dd2 than Dd2NM along the 5'-UTRs of the RH4 gene at both 44 and 24 h. Our data indicate that the failure of Dd2 to express the sialic acid-independent invasion receptor gene RH4 is associated with the epigenetic silencing mark H3K9 trimethylation present throughout the cycle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Upregulation of expression of the reticulocyte homology gene 4 in the Plasmodium falciparum clone Dd2 is associated with a switch in the erythrocyte invasion pathway.Mol Biochem Parasitol. 2006 Feb;145(2):205-15. doi: 10.1016/j.molbiopara.2005.10.004. Epub 2005 Oct 25. Mol Biochem Parasitol. 2006. PMID: 16289357
-
Recombinant Plasmodium falciparum reticulocyte homology protein 4 binds to erythrocytes and blocks invasion.Proc Natl Acad Sci U S A. 2007 Nov 6;104(45):17789-94. doi: 10.1073/pnas.0708772104. Epub 2007 Oct 30. Proc Natl Acad Sci U S A. 2007. PMID: 17971435 Free PMC article.
-
Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes.Science. 2005 Aug 26;309(5739):1384-7. doi: 10.1126/science.1115257. Science. 2005. PMID: 16123303
-
Plasmodium falciparum: epigenetic control of var gene regulation and disease.Subcell Biochem. 2013;61:659-82. doi: 10.1007/978-94-007-4525-4_28. Subcell Biochem. 2013. PMID: 23150271 Review.
-
The role of chromatin in Plasmodium gene expression.Cell Microbiol. 2012 Jun;14(6):819-28. doi: 10.1111/j.1462-5822.2012.01777.x. Epub 2012 Mar 28. Cell Microbiol. 2012. PMID: 22360617 Review.
Cited by
-
Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites.PLoS Genet. 2012;8(11):e1002992. doi: 10.1371/journal.pgen.1002992. Epub 2012 Nov 1. PLoS Genet. 2012. PMID: 23133397 Free PMC article.
-
Variation in Plasmodium falciparum erythrocyte invasion phenotypes and merozoite ligand gene expression across different populations in areas of malaria endemicity.Infect Immun. 2015 Jun;83(6):2575-82. doi: 10.1128/IAI.03009-14. Epub 2015 Apr 13. Infect Immun. 2015. PMID: 25870227 Free PMC article.
-
Heterochromatin de novo formation and maintenance in Plasmodium falciparum.PLoS Pathog. 2025 Jun 2;21(6):e1013137. doi: 10.1371/journal.ppat.1013137. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40455742 Free PMC article.
-
Epigenetic and Epitranscriptomic Gene Regulation in Plasmodium falciparum and How We Can Use It against Malaria.Genes (Basel). 2022 Sep 27;13(10):1734. doi: 10.3390/genes13101734. Genes (Basel). 2022. PMID: 36292619 Free PMC article. Review.
-
Stochastic expression of invasion genes in Plasmodium falciparum schizonts.Nat Commun. 2022 May 30;13(1):3004. doi: 10.1038/s41467-022-30605-z. Nat Commun. 2022. PMID: 35637187 Free PMC article.
References
-
- Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. - PubMed
-
- Gaur D, et al. Upregulation of expression of the reticulocyte homology gene 4 in the Plasmodium falciparum clone Dd2 is associated with a switch in the erythrocyte invasion pathway. Mol Biochem Parasitol. 2006;145:205–215. - PubMed
-
- Stubbs J, et al. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science. 2005;309:1384–1387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

