Detecting and understanding combinatorial mutation patterns responsible for HIV drug resistance
- PMID: 20080674
- PMCID: PMC2824344
- DOI: 10.1073/pnas.0907304107
Detecting and understanding combinatorial mutation patterns responsible for HIV drug resistance
Abstract
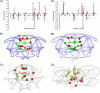
We propose a systematic approach for a better understanding of how HIV viruses employ various combinations of mutations to resist drug treatments, which is critical to developing new drugs and optimizing the use of existing drugs. By probabilistically modeling mutations in the HIV-1 protease or reverse transcriptase (RT) isolated from drug-treated patients, we present a statistical procedure that first detects mutation combinations associated with drug resistance and then infers detailed interaction structures of these mutations. The molecular basis of our statistical predictions is further studied by using molecular dynamics simulations and free energy calculations. We have demonstrated the usefulness of this systematic procedure on three HIV drugs, (Indinavir, Zidovudine, and Nevirapine), discovered unique interaction features between viral mutations induced by these drugs, and revealed the structural basis of such interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
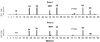

References
-
- Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol. 2007;25:1407–1410. - PubMed
-
- Lengauer T, Sing L. Bioinformatics-assisted anti-HIV therapy. Nat Rev Microbiol. 2006;4:790–797. - PubMed
-
- Johnson VA, et al. Update of the drug resistance mutations in HIV-1: Spring 2008. Top HIV Med. 2008;16:62–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

