Lack of evolvability in self-sustaining autocatalytic networks constraints metabolism-first scenarios for the origin of life
- PMID: 20080693
- PMCID: PMC2824406
- DOI: 10.1073/pnas.0912628107
Lack of evolvability in self-sustaining autocatalytic networks constraints metabolism-first scenarios for the origin of life
Abstract
A basic property of life is its capacity to experience Darwinian evolution. The replicator concept is at the core of genetics-first theories of the origin of life, which suggest that self-replicating oligonucleotides or their similar ancestors may have been the first "living" systems and may have led to the evolution of an RNA world. But problems with the nonenzymatic synthesis of biopolymers and the origin of template replication have spurred the alternative metabolism-first scenario, where self-reproducing and evolving proto-metabolic networks are assumed to have predated self-replicating genes. Recent theoretical work shows that "compositional genomes" (i.e., the counts of different molecular species in an assembly) are able to propagate compositional information and can provide a setup on which natural selection acts. Accordingly, if we stick to the notion of replicator as an entity that passes on its structure largely intact in successive replications, those macromolecular aggregates could be dubbed "ensemble replicators" (composomes) and quite different from the more familiar genes and memes. In sharp contrast with template-dependent replication dynamics, we demonstrate here that replication of compositional information is so inaccurate that fitter compositional genomes cannot be maintained by selection and, therefore, the system lacks evolvability (i.e., it cannot substantially depart from the asymptotic steady-state solution already built-in in the dynamical equations). We conclude that this fundamental limitation of ensemble replicators cautions against metabolism-first theories of the origin of life, although ancient metabolic systems could have provided a stable habitat within which polymer replicators later evolved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
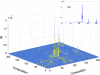
 in Eq. 6. GARD assemblies were characterized as 10-long vectors and distinguished by their initial composition of
in Eq. 6. GARD assemblies were characterized as 10-long vectors and distinguished by their initial composition of  molecules (
molecules ( ). The values for the forward and backward rate constants as described by Eq. 1 with
). The values for the forward and backward rate constants as described by Eq. 1 with  were
were  and
and  (i.e., we have neglected decay of assemblies), respectively. The elements of the
(i.e., we have neglected decay of assemblies), respectively. The elements of the  matrix for the catalytic enhancement factors were sampled from a log-normal distribution with parameters
matrix for the catalytic enhancement factors were sampled from a log-normal distribution with parameters  and
and  as in ref. . As a legacy, the elements in the
as in ref. . As a legacy, the elements in the  matrix (
matrix ( ) have mean 18,157.3 and variance
) have mean 18,157.3 and variance  , and some off-diagonal elements can be as large, or even larger, than the diagonal elements. The inset plot shows the particular distribution for row 94; that is, the values that lead to the increase in frequency of the most frequent composome
, and some off-diagonal elements can be as large, or even larger, than the diagonal elements. The inset plot shows the particular distribution for row 94; that is, the values that lead to the increase in frequency of the most frequent composome  (
( and
and  , which means that the outflow from these compositions to the equilibrium frequency of the leading composome is greater than the bona fide replication of the leading composome itself.
, which means that the outflow from these compositions to the equilibrium frequency of the leading composome is greater than the bona fide replication of the leading composome itself.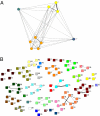
 matrices of molecular repertoires (A)
matrices of molecular repertoires (A)  and (B)
and (B)  . Nodes marked with the same color belong to the same quasicompartment. The widths of links correspond to the interaction strength. For the large
. Nodes marked with the same color belong to the same quasicompartment. The widths of links correspond to the interaction strength. For the large  matrix (B), only interactions stronger than
matrix (B), only interactions stronger than  are shown for simplicity. Quasicompartments are formed where the few large
are shown for simplicity. Quasicompartments are formed where the few large  values from a log-normal distribution fall. The figure was drawn using NetDraw (42).
values from a log-normal distribution fall. The figure was drawn using NetDraw (42).
 values (Eq. 2) along 54 generations derived from computer simulations of the GARD model using the
values (Eq. 2) along 54 generations derived from computer simulations of the GARD model using the  matrix in Fig. 2B. Only individuals before splitting are included in the analysis. Purple squares mark QSSs where three different composomes (A, B, and C) persist.
matrix in Fig. 2B. Only individuals before splitting are included in the analysis. Purple squares mark QSSs where three different composomes (A, B, and C) persist.
Similar articles
-
Primordial evolvability: Impasses and challenges.J Theor Biol. 2015 Sep 21;381:29-38. doi: 10.1016/j.jtbi.2015.06.047. Epub 2015 Jul 9. J Theor Biol. 2015. PMID: 26165453
-
Systems protobiology: origin of life in lipid catalytic networks.J R Soc Interface. 2018 Jul;15(144):20180159. doi: 10.1098/rsif.2018.0159. J R Soc Interface. 2018. PMID: 30045888 Free PMC article. Review.
-
The origin of replicators and reproducers.Philos Trans R Soc Lond B Biol Sci. 2006 Oct 29;361(1474):1761-76. doi: 10.1098/rstb.2006.1912. Philos Trans R Soc Lond B Biol Sci. 2006. PMID: 17008217 Free PMC article.
-
Origin of life in a digital microcosm.Philos Trans A Math Phys Eng Sci. 2017 Dec 28;375(2109):20160350. doi: 10.1098/rsta.2016.0350. Philos Trans A Math Phys Eng Sci. 2017. PMID: 29133448 Free PMC article.
-
Metabolically Coupled Replicator Systems: Overview of an RNA-world model concept of prebiotic evolution on mineral surfaces.J Theor Biol. 2015 Sep 21;381:39-54. doi: 10.1016/j.jtbi.2015.06.002. Epub 2015 Jun 15. J Theor Biol. 2015. PMID: 26087284 Review.
Cited by
-
Spontaneous chiral symmetry breaking in early molecular networks.Biol Direct. 2010 May 27;5:38. doi: 10.1186/1745-6150-5-38. Biol Direct. 2010. PMID: 20507625 Free PMC article.
-
Small-molecule autocatalysis drives compartment growth, competition and reproduction.Nat Chem. 2024 Jan;16(1):70-78. doi: 10.1038/s41557-023-01276-0. Epub 2023 Aug 7. Nat Chem. 2024. PMID: 37550391
-
Serpentinite and the dawn of life.Philos Trans R Soc Lond B Biol Sci. 2011 Oct 27;366(1580):2857-69. doi: 10.1098/rstb.2011.0129. Philos Trans R Soc Lond B Biol Sci. 2011. PMID: 21930576 Free PMC article.
-
The Informational Substrate of Chemical Evolution: Implications for Abiogenesis.Life (Basel). 2019 Aug 8;9(3):66. doi: 10.3390/life9030066. Life (Basel). 2019. PMID: 31398942 Free PMC article.
-
Multilevel selection as Bayesian inference, major transitions in individuality as structure learning.R Soc Open Sci. 2019 Aug 28;6(8):190202. doi: 10.1098/rsos.190202. eCollection 2019 Aug. R Soc Open Sci. 2019. PMID: 31598234 Free PMC article.
References
-
- Chen QW, Chen CL. The role of inorganic compounds in the prebiotic synthesis of organic molecules. Curr Org Chem. 2005;9:989–998.
-
- Orgel LE. Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol. 2004;39:99–123. - PubMed
-
- Anet FA. The place of metabolism in the origin of life. Curr Opin Chem Biol. 2004;8:654–659. - PubMed
-
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. - PubMed
-
- Kun Á, Santos M, Szathmáry E. Real ribozymes suggest a relaxed error threshold. Nat Genet. 2005;37:1008–1011. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

