Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer
- PMID: 20080697
- PMCID: PMC2836606
- DOI: 10.1073/pnas.0904497107
Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer
Erratum in
- Proc Natl Acad Sci U S A. 2010 Apr 27;107(17):8041
Abstract
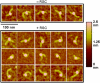
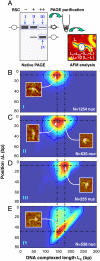
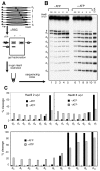
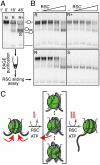
Chromatin remodelers are sophisticated nano-machines that are able to alter histone-DNA interactions and to mobilize nucleosomes. Neither the mechanism of their action nor the conformation of the remodeled nucleosomes are, however, yet well understood. We have studied the mechanism of Remodels Structure of Chromatin (RSC)-nucleosome mobilization by using high-resolution microscopy and biochemical techniques. Atomic force microscopy and electron cryomicroscopy (EC-M) analyses show that two types of products are generated during the RSC remodeling: (i) stable non-mobilized particles, termed remosomes that contain about 180 bp of DNA associated with the histone octamer and, (ii) mobilized particles located at the end of DNA. EC-M reveals that individual remosomes exhibit a distinct, variable, highly-irregular DNA trajectory. The use of the unique "one pot assays" for studying the accessibility of nucleosomal DNA towards restriction enzymes, DNase I footprinting and ExoIII mapping demonstrate that the histone-DNA interactions within the remosomes are strongly perturbed, particularly in the vicinity of the nucleosome dyad. The data suggest a two-step mechanism of RSC-nucleosome remodeling consisting of an initial formation of a remosome followed by mobilization. In agreement with this model, we show experimentally that the remosomes are intermediate products generated during the first step of the remodeling reaction that are further efficiently mobilized by RSC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans- displacement of the histone octamer. Cell. 1999;97(7):843–852. - PubMed
-
- Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2003;303(5656):343–348. - PubMed
-
- Bao Y, Shen X. SnapShot: chromatin remodeling complexes. Cell. 2007;129(3):632. - PubMed
-
- Cairns BR, et al. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87(7):1249–1260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

