Control of myosin-I force sensing by alternative splicing
- PMID: 20080738
- PMCID: PMC2818919
- DOI: 10.1073/pnas.0911426107
Control of myosin-I force sensing by alternative splicing
Abstract
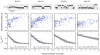
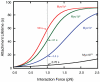
Myosin-Is are molecular motors that link cellular membranes to the actin cytoskeleton, where they play roles in mechano-signal transduction and membrane trafficking. Some myosin-Is are proposed to act as force sensors, dynamically modulating their motile properties in response to changes in tension. In this study, we examined force sensing by the widely expressed myosin-I isoform, myo1b, which is alternatively spliced in its light chain binding domain (LCBD), yielding proteins with lever arms of different lengths. We found the actin-detachment kinetics of the splice isoforms to be extraordinarily tension-sensitive, with the magnitude of tension sensitivity to be related to LCBD splicing. Thus, in addition to regulating step-size, motility rates, and myosin activation, the LCBD is a key regulator of force sensing. We also found that myo1b is substantially more tension-sensitive than other myosins with similar length lever arms, indicating that different myosins have different tension-sensitive transitions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

