Genomic scale analysis of racial impact on response to IFN-alpha
- PMID: 20080756
- PMCID: PMC2818915
- DOI: 10.1073/pnas.0913491107
Genomic scale analysis of racial impact on response to IFN-alpha
Abstract
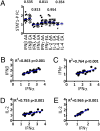
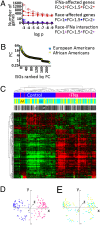
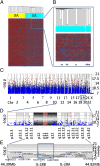
Limited responsiveness to IFN-alpha in hepatitis C virus (HCV)-infected African-Americans compared to European Americans (AAs vs. EAs) hinders the management of HCV. Here, we studied healthy non-HCV-infected AA and EA subjects to test whether immune cell response to IFN-alpha is determined directly by race. We compared baseline and IFN-alpha-induced signal transducer and activator of transcription (STAT)-1, STAT-2, STAT-3, STAT-4, and STAT-5 protein and phosphorylation levels in purified T cells, global transcription, and a genomewide single-nucleotide polymorphism (SNP) profile of healthy AA and EA blood donors. In contrast to HCV-infected individuals, healthy AAs displayed no evidence of reduced STAT activation or IFN-alpha-stimulated gene expression compared to EAs. Although >200 genes reacted to IFN-alpha treatment, race had no impact on any of them. The only gene differentially expressed by the two races (NUDT3, P < 10(-7)) was not affected by IFN-alpha and bears no known relationship to IFN-alpha signaling or HCV pathogenesis. Genomewide analysis confirmed the self-proclaimed racial attribution of most donors, and numerous race-associated SNPs were identified within loci involved in IFN-alpha signaling, although they clearly did not affect responsiveness in the absence of HCV. We conclude that racial differences observed in HCV-infected patients in the responsiveness to IFN-alpha are unrelated to inherent racial differences in IFN-alpha signaling and more likely due to polymorphisms affecting the hosts' response to HCV, which in turn may lead to a distinct disease pathophysiology responsible for altered IFN signaling and treatment response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Reddy KR, et al. Racial differences in responses to therapy with interferon in chronic hepatitis C. Consensus Interferon. Study Group. Hepatology. 1999;30:787–793. - PubMed
-
- He XS, et al. Global transcriptional response to interferon is a determinant of HCV treatment outcome and is modified by race. Hepatology. 2006;44:352–359. - PubMed
-
- Layden-Almer JE, Ribeiro RM, Wiley T, Perelson AS, Layden TJ. Viral dynamics and response differences in HCV-infected African American and white patients treated with IFN and ribavirin. Hepatology. 2003;37:1343–1350. - PubMed
-
- Muir AJ, Bornstein JD, Killenberg PG Atlantic Coast Hepatitis Treatment Group. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med. 2004;350:2265–2271. - PubMed
-
- Hepburn MJ, Hepburn LM, Cantu NS, Lapeer MG, Lawitz EJ. Differences in treatment outcome for hepatitis C among ethnic groups. Am J Med. 2004;117:163–168. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

