The induction of folding cooperativity by ligand binding drives the allosteric response of tetracycline repressor
- PMID: 20080791
- PMCID: PMC2799725
- DOI: 10.1073/pnas.0911566106
The induction of folding cooperativity by ligand binding drives the allosteric response of tetracycline repressor
Abstract
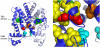
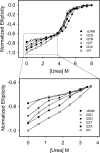
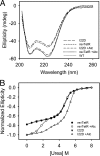
Tetracycline (Tc) repressor (TetR) undergoes an allosteric transition upon interaction with the antibiotic, Tc, that abrogates its ability to specifically bind its operator DNA. In this work, by performing equilibrium protein unfolding experiments on wild-type TetR and mutants displaying altered allosteric responses, we have delineated a model to explain TetR allostery. In the absence of Tc, we show that the DNA-binding domains of this homodimeric protein are relatively flexible and unfold independently of the Tc binding/dimerization (TBD) domains. Once Tc is bound, however, the unfolding of the DNA binding domains becomes coupled to the TBD domains, leading to a large increase in DNA-binding domain stability. Noninducible TetR mutants display considerably less interdomain folding cooperativity upon binding to Tc. We conclude that the thermodynamic coupling of the TetR domains caused by Tc binding and the resulting rigidification of the DNA-binding domains into a conformation that is incompatible with DNA binding are the fundamental factors leading to the allosteric response in TetR. This allosteric mechanism can account for properties of the whole TetR family of repressors and may explain the functioning and evolution of other allosteric systems. Our model contrasts with the prevalent view that TetR populates two distinct conformations and that Tc causes a switch between these defined conformations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
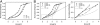

Comment in
-
Regulating transcription regulators via allostery and flexibility.Proc Natl Acad Sci U S A. 2009 Dec 29;106(52):22035-6. doi: 10.1073/pnas.0912300107. Epub 2009 Dec 23. Proc Natl Acad Sci U S A. 2009. PMID: 20080782 Free PMC article. Review. No abstract available.
Similar articles
-
Two-way interdomain signal transduction in tetracycline repressor.J Mol Biol. 2006 Aug 11;361(2):382-9. doi: 10.1016/j.jmb.2006.06.035. Epub 2006 Jun 30. J Mol Biol. 2006. PMID: 16844141
-
Tet repressor induction by tetracycline: a molecular dynamics, continuum electrostatics, and crystallographic study.J Mol Biol. 2008 May 9;378(4):898-912. doi: 10.1016/j.jmb.2008.03.022. Epub 2008 Mar 19. J Mol Biol. 2008. PMID: 18395746
-
Tet repressor mutants with altered effector binding and allostery.FEBS J. 2005 Sep;272(17):4487-96. doi: 10.1111/j.1742-4658.2005.04868.x. FEBS J. 2005. PMID: 16128817
-
Regulating transcription regulators via allostery and flexibility.Proc Natl Acad Sci U S A. 2009 Dec 29;106(52):22035-6. doi: 10.1073/pnas.0912300107. Epub 2009 Dec 23. Proc Natl Acad Sci U S A. 2009. PMID: 20080782 Free PMC article. Review. No abstract available.
-
The whole lactose repressor.Science. 1996 Mar 1;271(5253):1245-6. doi: 10.1126/science.271.5253.1245. Science. 1996. PMID: 8638104 Review. No abstract available.
Cited by
-
Induced fit, conformational selection and independent dynamic segments: an extended view of binding events.Trends Biochem Sci. 2010 Oct;35(10):539-46. doi: 10.1016/j.tibs.2010.04.009. Epub 2010 Jun 11. Trends Biochem Sci. 2010. PMID: 20541943 Free PMC article.
-
The "violin model": Looking at community networks for dynamic allostery.J Chem Phys. 2023 Feb 28;158(8):081001. doi: 10.1063/5.0138175. J Chem Phys. 2023. PMID: 36859094 Free PMC article. Review.
-
The ensemble nature of allostery.Nature. 2014 Apr 17;508(7496):331-9. doi: 10.1038/nature13001. Nature. 2014. PMID: 24740064 Free PMC article. Review.
-
Structural and functional characterization of a ketosteroid transcriptional regulator of Mycobacterium tuberculosis.J Biol Chem. 2015 Jan 9;290(2):872-82. doi: 10.1074/jbc.M114.607481. Epub 2014 Nov 18. J Biol Chem. 2015. PMID: 25406313 Free PMC article.
-
Molecular dynamics simulation unveils the conformational flexibility of the interdomain linker in the bacterial transcriptional regulator GabR from Bacillus subtilis bound to pyridoxal 5'-phosphate.PLoS One. 2017 Dec 18;12(12):e0189270. doi: 10.1371/journal.pone.0189270. eCollection 2017. PLoS One. 2017. PMID: 29253008 Free PMC article.
References
-
- Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr Opin Struct Biol. 2006;16:102–108. - PubMed
-
- Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. - PubMed
-
- Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. - PubMed
-
- Kern D, Zuiderweg ER. The role of dynamics in allosteric regulation. Curr Opin Struct Biol. 2003;13:748–757. - PubMed
-
- Luque I, Leavitt SA, Freire E. The linkage between protein folding and functional cooperativity: Two sides of the same coin? Annu Rev Biophys Biomol Struct. 2002;31:235–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

