Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65
- PMID: 20080798
- PMCID: PMC2806709
- DOI: 10.1073/pnas.0912493107
Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65
Abstract
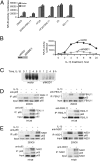
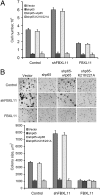
NF-kappaB, a central coordinator of immune and inflammatory responses, must be tightly regulated. We describe a NF-kappaB regulatory pathway that is driven by reversible lysine methylation of the p65 subunit, carried out by a lysine methylase, the nuclear receptor-binding SET domain-containing protein 1 (NSD1), and a lysine demethylase, F-box and leucine-rich repeat protein 11 (FBXL11). Overexpression of FBXL11 inhibits NF-kappaB activity, and a high level of NSD1 activates NF-kappaB and reverses the inhibitory effect of FBXL11, whereas reduced expression of NSD1 decreases NF-kappaB activation. The targets are K218 and K221 of p65, which are methylated in cells with activated NF-kappaB. Overexpression of FBXL11 slowed the growth of HT29 cancer cells, whereas shRNA-mediated knockdown had the opposite effect, and these phenotypes were dependent on K218/K221 methylation. In mouse embryo fibroblasts, the activation of most p65-dependent genes relied on K218/K221 methylation. Importantly, expression of the FBXL11 gene is driven by NF-kappaB, revealing a negative regulatory feedback loop. We conclude that reversible lysine methylation of NF-kappaB is an important element in the complex regulation of this key transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Role of lysine methylation of NF-κB in differential gene regulation.Proc Natl Acad Sci U S A. 2013 Aug 13;110(33):13510-5. doi: 10.1073/pnas.1311770110. Epub 2013 Jul 31. Proc Natl Acad Sci U S A. 2013. PMID: 23904479 Free PMC article.
-
Nuclear receptor binding SET domain protein 1 promotes epithelial-mesenchymal transition in paclitaxel-resistant breast cancer cells via regulating nuclear factor kappa B and F-box and leucine-rich repeat protein 11.Bioengineered. 2021 Dec;12(2):11506-11519. doi: 10.1080/21655979.2021.2009963. Bioengineered. 2021. PMID: 34905470 Free PMC article.
-
Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit.EMBO J. 2009 Apr 22;28(8):1055-66. doi: 10.1038/emboj.2009.55. Epub 2009 Mar 5. EMBO J. 2009. PMID: 19262565 Free PMC article.
-
The NSD family of protein methyltransferases in human cancer.Epigenomics. 2015 Aug;7(5):863-74. doi: 10.2217/epi.15.32. Epub 2015 May 5. Epigenomics. 2015. PMID: 25942451 Review.
-
The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection.Cell Cycle. 2007 Jun 1;6(11):1324-8. doi: 10.4161/cc.6.11.4269. Epub 2007 Jun 14. Cell Cycle. 2007. PMID: 17568193 Review.
Cited by
-
Post-translational modifications of p65: state of the art.Front Cell Dev Biol. 2024 Jul 10;12:1417502. doi: 10.3389/fcell.2024.1417502. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39050887 Free PMC article. Review.
-
Methods to detect NF-κB acetylation and methylation.Methods Mol Biol. 2015;1280:395-409. doi: 10.1007/978-1-4939-2422-6_24. Methods Mol Biol. 2015. PMID: 25736763 Free PMC article.
-
Understanding the language of Lys36 methylation at histone H3.Nat Rev Mol Cell Biol. 2012 Jan 23;13(2):115-26. doi: 10.1038/nrm3274. Nat Rev Mol Cell Biol. 2012. PMID: 22266761 Free PMC article. Review.
-
Lysine methylation signaling of non-histone proteins in the nucleus.Cell Mol Life Sci. 2019 Aug;76(15):2873-2883. doi: 10.1007/s00018-019-03142-0. Epub 2019 May 23. Cell Mol Life Sci. 2019. PMID: 31123776 Free PMC article. Review.
-
Inhibition of LSD1 phosphorylation alleviates colitis symptoms induced by dextran sulfate sodium.BMB Rep. 2020 Jul;53(7):385-390. doi: 10.5483/BMBRep.2020.53.7.298. BMB Rep. 2020. PMID: 32317082 Free PMC article.
References
-
- Karin M, Yamamoto Y, Wang QM. The IKK NF-κB system: A treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26. - PubMed
-
- Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. - PubMed
-
- Lu T, Sathe SS, Swiatkowski SM, Hampole CV, Stark GR. Secretion of cytokines and growth factors as a general cause of constitutive NFκB activation in cancer. Oncogene. 2004;23:2138–2145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

