Biofilm formation by and multicellular behavior of Escherichia coli O55:H7, an atypical enteropathogenic strain
- PMID: 20080991
- PMCID: PMC2832381
- DOI: 10.1128/AEM.01395-09
Biofilm formation by and multicellular behavior of Escherichia coli O55:H7, an atypical enteropathogenic strain
Abstract
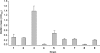
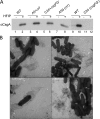
Enteropathogenic Escherichia coli (EPEC) is an important causal agent of diarrheal illness throughout the world. Nevertheless, researchers have only recently begun to explore its capacity to form biofilms. Strain O55:H7 (DMS9) is a clinical isolate belonging to the atypical EPEC (aEPEC) group, which displays a high degree of genetic relatedness to enterohemorrhagic E. coli. Strain DMS9 formed a robust biofilm on an abiotic surface at 26 degrees C, but not at 37 degrees C. It also formed a dense pellicle at the air-liquid interface and developed a red, rough, and dry (RDAR) morphotype on Congo red agar. Unlike a previously described E. coli O157:H7 strain, the aEPEC strain seems to express cellulose. Transposon mutagenesis was used to identify biofilm-deficient mutants. One of the mutants was inactivated in the csgFG genes, required for assembly and secretion of curli fimbriae, while a second mutant had a mutation in crl, a thermosensitive global regulator that modulates sigma(S) activity and downstream expression of curli and cellulose. The two mutants were deficient in their biofilm formation capabilities and did not form a pellicle at the air-liquid interface. Unlike in Salmonella, the csgFG mutant in aEPEC completely lost the RDAR phenotype, while the crl mutant displayed a unique RDAR "pizza"-like morphotype. Genetic complementation of the two mutants resulted in restoration of the wild-type phenotype. This report is the first to describe and analyze a multicellular behavior in aEPEC and support a major role for curli and the crl regulator in biofilm development at low temperatures corresponding to the nonmammalian host environment.
Figures


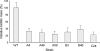

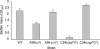



Similar articles
-
Phenotypic and genotypic characteristics associated with biofilm formation in clinical isolates of atypical enteropathogenic Escherichia coli (aEPEC) strains.BMC Microbiol. 2014 Jul 10;14:184. doi: 10.1186/1471-2180-14-184. BMC Microbiol. 2014. PMID: 25012525 Free PMC article.
-
Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli.Environ Microbiol. 2009 Apr;11(4):992-1006. doi: 10.1111/j.1462-2920.2008.01824.x. Epub 2009 Nov 14. Environ Microbiol. 2009. PMID: 19187284 Free PMC article.
-
Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents.Appl Environ Microbiol. 2006 Apr;72(4):2564-72. doi: 10.1128/AEM.72.4.2564-2572.2006. Appl Environ Microbiol. 2006. PMID: 16597958 Free PMC article.
-
Characterization of biofilm-forming capacity and resistance to sanitizers of a range of E. coli O26 pathotypes from clinical cases and cattle in Australia.BMC Microbiol. 2018 May 8;18(1):41. doi: 10.1186/s12866-018-1182-z. BMC Microbiol. 2018. PMID: 29739319 Free PMC article.
-
Identification of novel biofilm genes in avian pathogenic Escherichia coli by Tn5 transposon mutant library.World J Microbiol Biotechnol. 2022 Jun 11;38(8):130. doi: 10.1007/s11274-022-03314-4. World J Microbiol Biotechnol. 2022. PMID: 35688968 Review.
Cited by
-
Metal-Adapted Bacteria Isolated From Wastewaters Produce Biofilms by Expressing Proteinaceous Curli Fimbriae and Cellulose Nanofibers.Front Microbiol. 2018 Jun 25;9:1334. doi: 10.3389/fmicb.2018.01334. eCollection 2018. Front Microbiol. 2018. PMID: 29988579 Free PMC article.
-
Biofilm modifies expression of ribonucleotide reductase genes in Escherichia coli.PLoS One. 2012;7(9):e46350. doi: 10.1371/journal.pone.0046350. Epub 2012 Sep 26. PLoS One. 2012. PMID: 23050019 Free PMC article.
-
Fimbrial adhesins produced by atypical enteropathogenic Escherichia coli strains.Appl Environ Microbiol. 2011 Dec;77(23):8391-9. doi: 10.1128/AEM.05376-11. Epub 2011 Sep 16. Appl Environ Microbiol. 2011. PMID: 21926222 Free PMC article.
-
Bacterial amyloids.Methods Mol Biol. 2012;849:303-20. doi: 10.1007/978-1-61779-551-0_21. Methods Mol Biol. 2012. PMID: 22528099 Free PMC article.
-
Dissection of the role of pili and type 2 and 3 secretion systems in adherence and biofilm formation of an atypical enteropathogenic Escherichia coli strain.Infect Immun. 2013 Oct;81(10):3793-802. doi: 10.1128/IAI.00620-13. Epub 2013 Jul 29. Infect Immun. 2013. PMID: 23897608 Free PMC article.
References
-
- Aidar-Ugrinovich, L., J. Blanco, M. Blanco, J. E. Blanco, L. Leomil, G. Dahbi, A. Mora, D. L. Onuma, W. D. Silveira, and A. F. P. de Castro. 2007. Serotypes, virulence genes, and intimin types of Shiga toxin-producing Escherichia coli (STEC) and enteropathogenic E. coli (EPEC) isolated from calves in Sao Paulo, Brazil. Int. J. Food Microbiol. 115:297-306. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Arnqvist, A., A. Olsen, J. Pfeifer, D. G. Russell, and S. Normark. 1992. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol. Microbiol. 6:2443-2452. - PubMed
-
- Austin, J. W., G. Sanders, W. Kay, and S. Collinson. 1998. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162:295. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

