Comparison of two optical-density-based methods and a plate count method for estimation of growth parameters of Bacillus cereus
- PMID: 20081006
- PMCID: PMC2832364
- DOI: 10.1128/AEM.02336-09
Comparison of two optical-density-based methods and a plate count method for estimation of growth parameters of Bacillus cereus
Abstract
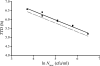
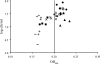
Quantitative microbiological models predicting proliferation of microorganisms relevant for food safety and/or food stability are useful tools to limit the need for generation of biological data through challenge testing and shelf-life testing. The use of these models requires quick and reliable methods for the generation of growth data and estimation of growth parameters. Growth parameter estimation can be achieved using methods based on plate counting and methods based on measuring the optical density. This research compares the plate count method with two optical density methods, namely, the 2-fold dilution (2FD) method and the relative rate to detection (RRD) method. For model organism Bacillus cereus F4810/72, the plate count method and both optical density methods gave comparable estimates for key growth parameters. Values for the maximum specific growth rate (mu(max)) derived by the 2FD method and by the RRD method were of the same order of magnitude, but some marked differences between the two approaches were apparent. Whereas the 2FD method allowed the derivation of values for lag time (lambda) from the data, this was not possible with the RRD method. However, the RRD method gave many more data points per experiment and also gave more data points close to the growth boundary. This research shows that all three proposed methods can be used for parameter estimation but that the choice of method depends on the objectives of the research.
Figures
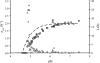


Similar articles
-
Comparison of Primary Models to Predict Microbial Growth by the Plate Count and Absorbance Methods.Biomed Res Int. 2015;2015:365025. doi: 10.1155/2015/365025. Epub 2015 Oct 11. Biomed Res Int. 2015. PMID: 26539483 Free PMC article.
-
Modelling Bacillus cereus growth.Int J Food Microbiol. 1997 Sep 16;38(2-3):229-34. doi: 10.1016/s0168-1605(97)00110-4. Int J Food Microbiol. 1997. PMID: 9506288
-
Growth kinetics of listeria isolated from salmon and salmon processing environment: single strains versus cocktails.J Food Prot. 2012 Jul;75(7):1227-35. doi: 10.4315/0362-028X.JFP-11-536. J Food Prot. 2012. PMID: 22980005
-
[Comparison of direct colony count methods and the MPN-method for quantitative detection of Listeria in model and field conditions].Berl Munch Tierarztl Wochenschr. 2001 Nov-Dec;114(11-12):453-64. Berl Munch Tierarztl Wochenschr. 2001. PMID: 11766274 Review. German.
-
Comparison of microbiological and physicochemical methods for enumeration of microorganisms.Postepy Hig Med Dosw (Online). 2014 Jan 2;68:1392-6. doi: 10.5604/17322693.1130086. Postepy Hig Med Dosw (Online). 2014. PMID: 25531702 Review.
Cited by
-
Quantifying Variability in Growth and Thermal Inactivation Kinetics of Lactobacillus plantarum.Appl Environ Microbiol. 2016 Jul 29;82(16):4896-908. doi: 10.1128/AEM.00277-16. Print 2016 Aug 15. Appl Environ Microbiol. 2016. PMID: 27260362 Free PMC article.
-
Controlling Microbial Safety Challenges of Meat Using High Voltage Atmospheric Cold Plasma.Front Microbiol. 2016 Jun 22;7:977. doi: 10.3389/fmicb.2016.00977. eCollection 2016. Front Microbiol. 2016. PMID: 27446018 Free PMC article.
-
Large plasmidome of dairy Lactococcus lactis subsp. lactis biovar diacetylactis FM03P encodes technological functions and appears highly unstable.BMC Genomics. 2018 Aug 17;19(1):620. doi: 10.1186/s12864-018-5005-2. BMC Genomics. 2018. PMID: 30119641 Free PMC article.
-
Estimation of Mycobacterium avium subsp. paratuberculosis growth parameters: strain characterization and comparison of methods.Appl Environ Microbiol. 2011 Dec;77(24):8615-24. doi: 10.1128/AEM.05818-11. Epub 2011 Oct 14. Appl Environ Microbiol. 2011. PMID: 22003015 Free PMC article.
-
Bacterial-like nitric oxide synthase in the haloalkaliphilic archaeon Natronomonas pharaonis.Microbiologyopen. 2020 Nov;9(11):e1124. doi: 10.1002/mbo3.1124. Epub 2020 Oct 14. Microbiologyopen. 2020. PMID: 33306280 Free PMC article.
References
-
- Adams, M. R., and M. O. Moss. 2000. Food microbiology, 2nd ed. The Royal Society of Chemistry, Cambridge, United Kingdom.
-
- Bidlas, E., T. Du, and R. J. W. Lambert. 2008. An explanation for the effect of inoculum size on MIC and the growth/no growth interface. Int. J. Food Microbiol. 126:140-152. - PubMed
-
- Browne, N., and B. C. A. Dowds. 2002. Acid stress in the food pathogen Bacillus cereus. J. Appl. Microbiol. 92:404-414. - PubMed
-
- Cuppers, H. G. A. M., and J. P. P. M. Smelt. 1993. Time to turbidity measurement as a tool for modeling spoilage by Lactobacillus. J. Ind. Microbiol. Biotechnol. 12:168-171.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

