Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35
- PMID: 20081868
- PMCID: PMC2872155
- DOI: 10.1038/nsmb.1765
Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35
Abstract
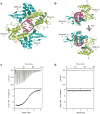
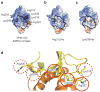
Ebola viral protein 35 (VP35), encoded by the highly pathogenic Ebola virus, facilitates host immune evasion by antagonizing antiviral signaling pathways, including those initiated by RIG-I-like receptors. Here we report the crystal structure of the Ebola VP35 interferon inhibitory domain (IID) bound to short double-stranded RNA (dsRNA), which together with in vivo results reveals how VP35-dsRNA interactions contribute to immune evasion. Conserved basic residues in VP35 IID recognize the dsRNA backbone, whereas the dsRNA blunt ends are 'end-capped' by a pocket of hydrophobic residues that mimic RIG-I-like receptor recognition of blunt-end dsRNA. Residues critical for RNA binding are also important for interferon inhibition in vivo but not for viral polymerase cofactor function of VP35. These results suggest that simultaneous recognition of dsRNA backbone and blunt ends provides a mechanism by which Ebola VP35 antagonizes host dsRNA sensors and immune responses.
Figures
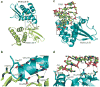
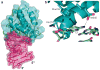
References
-
- Bosio CM, et al. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J Infect Dis. 2003;188:1630–8. - PubMed
-
- Mahanty S, et al. Protection from lethal infection is determined by innate immune responses in a mouse model of Ebola virus infection. Virology. 2003;312:415–24. - PubMed
-
- Bray M, Geisbert TW. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol. 2005;37:1560–6. - PubMed
-
- Baize S, et al. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5:423–6. - PubMed
-
- Feldmann H, Wahl-Jensen V, Jones SM, Stroher U. Ebola virus ecology: a continuing mystery. Trends Microbiol. 2004;12:433–7. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R01GM053163/GM/NIGMS NIH HHS/United States
- R01 GM053163/GM/NIGMS NIH HHS/United States
- R01AI059536/AI/NIAID NIH HHS/United States
- U54 AI057160/AI/NIAID NIH HHS/United States
- R01 NS010546/NS/NINDS NIH HHS/United States
- R01 AI081914/AI/NIAID NIH HHS/United States
- R01NS010546/NS/NINDS NIH HHS/United States
- F32 AI084324/AI/NIAID NIH HHS/United States
- R01AI081914/AI/NIAID NIH HHS/United States
- U54AI057160/AI/NIAID NIH HHS/United States
- R01 AI059536/AI/NIAID NIH HHS/United States
- AI057158/AI/NIAID NIH HHS/United States
- 1F32AI084324/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

