Characterization of the quinolone resistant determining regions in clinical isolates of pneumococci collected in Canada
- PMID: 20082699
- PMCID: PMC2823643
- DOI: 10.1186/1476-0711-9-3
Characterization of the quinolone resistant determining regions in clinical isolates of pneumococci collected in Canada
Abstract
Background: The objective of this study was to examine Streptococcus pneumoniae isolates collected from a longitudinal surveillance program in order to determine their susceptibility to currently used fluoroquinolones and of the frequency and type of mutations in the quinolone-resistant determining regions (QRDRs) of their parC and gyrA genes.
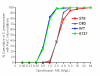
Methods: The Canadian Bacterial Surveillance Network has been collecting clinical isolates of S. pneumoniae from across Canada since 1988. Broth microdilution susceptibility testing was carried out according to the Clinical and Laboratory Standards Institute guidelines. The QRDRs of the parC and gyrA genes were sequenced for all isolates with ciprofloxacin MIC > or = 4 mg/L, and a large representative sample of isolates (N = 4,243) with MIC < or = 2 mg/L.
Results: A total of 4,798 out of 30,111 isolates collected from 1988, and 1993 to 2007 were studied. Of those isolates that were successfully sequenced, 184 out of 1,032 with mutations in parC only, 11 out of 30 with mutations in gyrA only, and 292 out of 298 with mutations in parC and gyrA were considered resistant to ciprofloxacin (MIC > or = 4 mg/L). The most common substitutions in the parC were at positions 137 (n = 722), 79 (n = 209), and 83 (n = 56), of which substitutions at positions 79 and 83 were associated with 4-fold increase in MIC to ciprofloxacin, whereas substitutions at position 137 had minimal effect on the ciprofloxacin MIC. A total of 400 out of 622 isolates with Lys-137 parC mutation belonged to serotypes 1, 12, 31, 7A, 9V, 9N and 9L, whereas only 49 out of 3064 isolates with no mutations belonged to these serotypes. Twenty-one out of 30 isolates with substitutions at position 81 of the gyrA gene had an increased MIC to ciprofloxacin. Finally, we found that isolates with mutations in both parC and gyrA were significantly associated with increased MIC to fluoroquinolones.
Conclusions: Not all mutations, most frequently Lys-137, found in the QRDRs of the parC gene of S. pneumoniae is associated with an increased MIC to fluoroquinolones. The high prevalence of Lys-137 appears to be due to its frequent occurrence in common serotypes.
Figures

Similar articles
-
Fluoroquinolone resistance in Streptococcus pneumoniae in United States since 1994-1995.Antimicrob Agents Chemother. 2002 Mar;46(3):680-8. doi: 10.1128/AAC.46.3.680-688.2002. Antimicrob Agents Chemother. 2002. PMID: 11850248 Free PMC article.
-
Are fluoroquinolone-susceptible isolates of Streptococcus pneumoniae really susceptible? A comparison of resistance mechanisms in Canadian isolates from 1997 and 2003.J Antimicrob Chemother. 2005 Oct;56(4):769-72. doi: 10.1093/jac/dki315. Epub 2005 Aug 26. J Antimicrob Chemother. 2005. PMID: 16126779
-
In vitro activity of sitafloxacin against clinical strains of Streptococcus pneumoniae with defined amino acid substitutions in QRDRs of gyrase A and topoisomerase IV.J Antimicrob Chemother. 2006 Dec;58(6):1279-82. doi: 10.1093/jac/dkl427. Epub 2006 Oct 20. J Antimicrob Chemother. 2006. PMID: 17056610
-
Mutant prevention concentrations for single-step fluoroquinolone-resistant mutants of wild-type, efflux-positive, or ParC or GyrA mutation-containing Streptococcus pneumoniae isolates.Antimicrob Agents Chemother. 2004 Oct;48(10):3954-8. doi: 10.1128/AAC.48.10.3954-3958.2004. Antimicrob Agents Chemother. 2004. PMID: 15388458 Free PMC article.
-
Novel Ser79Leu and Ser81Ile substitutions in the quinolone resistance-determining regions of ParC topoisomerase IV and GyrA DNA gyrase subunits from recent fluoroquinolone-resistant Streptococcus pneumoniae clinical isolates.Antimicrob Agents Chemother. 2005 Jun;49(6):2479-86. doi: 10.1128/AAC.49.6.2479-2486.2005. Antimicrob Agents Chemother. 2005. PMID: 15917550 Free PMC article.
Cited by
-
Association of mutation patterns in GyrA and ParC genes with quinolone resistance levels in lactic acid bacteria.J Antibiot (Tokyo). 2015 Feb;68(2):81-7. doi: 10.1038/ja.2014.113. Epub 2014 Sep 10. J Antibiot (Tokyo). 2015. PMID: 25204345
-
Susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada.Antimicrob Agents Chemother. 2011 Aug;55(8):3703-8. doi: 10.1128/AAC.00237-11. Epub 2011 May 31. Antimicrob Agents Chemother. 2011. PMID: 21628545 Free PMC article.
-
Molecular Analysis of Rising Fluoroquinolone Resistance in Belgian Non-Invasive Streptococcus pneumoniae Isolates (1995-2014).PLoS One. 2016 May 26;11(5):e0154816. doi: 10.1371/journal.pone.0154816. eCollection 2016. PLoS One. 2016. PMID: 27227336 Free PMC article.
-
Control of fluoroquinolone resistance through successful regulation, Australia.Emerg Infect Dis. 2012 Sep;18(9):1453-60. doi: 10.3201/eid1809.111515. Emerg Infect Dis. 2012. PMID: 22932272 Free PMC article.
-
Molecular Investigation of Quinolone Resistance of Quinolone Resistance-Determining Region in Streptococcus pneumoniae Strains Isolated from Iran Using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism Method.Osong Public Health Res Perspect. 2014 Oct;5(5):245-50. doi: 10.1016/j.phrp.2014.08.010. Epub 2014 Sep 6. Osong Public Health Res Perspect. 2014. PMID: 25389509 Free PMC article.
References
-
- Musher DM. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin Infect Dis. 1992;14(4):801–807. - PubMed
-
- Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM Jr, Musher DM, Niederman MS, Torres A, Whitney CG. Infectious Diseases Society of America; American Thoracic Society. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–72. doi: 10.1086/511159. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

