Properties of Escherichia coli EF-Tu mutants designed for fluorescence resonance energy transfer from tRNA molecules
- PMID: 20083494
- PMCID: PMC2816605
- DOI: 10.1093/protein/gzp079
Properties of Escherichia coli EF-Tu mutants designed for fluorescence resonance energy transfer from tRNA molecules
Abstract
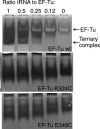
Here we describe the design, preparation and characterization of 10 EF-Tu mutants of potential utility for the study of Escherichia coli elongation factor Tu (EF-Tu) interaction with tRNA by a fluorescence resonance energy transfer assay. Each mutant contains a single cysteine residue at positions in EF-Tu that are proximal to tRNA sites within the aminoacyl-tRNA.EF-Tu.GTP ternary complex that have previously been labeled with fluorophores. These positions fall in the 323-326 and 344-348 regions of EF-Tu, and at the C terminus. The EF-Tus were isolated as N-terminal fusions to glutathione S-transferase (GST), which was cleaved to yield intact EF-Tus. The mutant EF-Tus were tested for binding to GDP, binding to tRNA in gel retardation and protection assays, and activity in poly-U translation in vitro. The results indicate that at least three EF-Tu mutants, K324C, G325C and E348C, are suitable for further studies. Remarkably, GST fusions that were not cleaved were also active in the various assays, despite the N-terminal fusion.
Figures
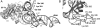
References
-
- Anborgh P.H., Parmeggiani A., Jonak J. Eur. J. Biochem. FEBS. 1992;208:251–257. - PubMed
-
- Andersen C., Wiborg O. Eur. J. Biochem. FEBS. 1994;220:739–744. - PubMed
-
- Blechschmidt B., Shirokov V., Sprinzl M. Eur. J. Biochem. FEBS. 1994;219:65–71. - PubMed
-
- Boon K., Vijgenboom E., Madsen L.V., Talens A., Kraal B., Bosch L. Eur. J. Biochem. FEBS. 1992;210:177–183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

