Flavonoid phytoalexin-dependent resistance to anthracnose leaf blight requires a functional yellow seed1 in Sorghum bicolor
- PMID: 20083611
- PMCID: PMC2865927
- DOI: 10.1534/genetics.109.111831
Flavonoid phytoalexin-dependent resistance to anthracnose leaf blight requires a functional yellow seed1 in Sorghum bicolor
Abstract
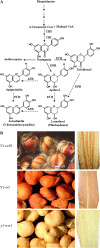
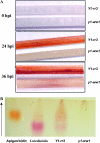
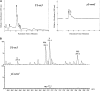
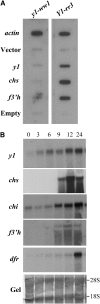
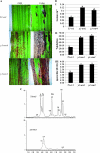
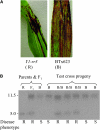
In Sorghum bicolor, a group of phytoalexins are induced at the site of infection by Colletotrichum sublineolum, the anthracnose fungus. These compounds, classified as 3-deoxyanthocyanidins, have structural similarities to the precursors of phlobaphenes. Sorghum yellow seed1 (y1) encodes a MYB transcription factor that regulates phlobaphene biosynthesis. Using the candystripe1 transposon mutagenesis system in sorghum, we have isolated functional revertants as well as loss-of-function alleles of y1. These near-isogenic lines of sorghum show that, compared to functionally revertant alleles, loss of y1 lines do not accumulate phlobaphenes. Molecular characterization of two null y1 alleles shows a partial internal deletion in the y1 sequence. These null alleles, designated as y1-ww1 and y1-ww4, do not accumulate 3-deoxyanthocyanidins when challenged with the nonpathogenic fungus Cochliobolus heterostrophus. Further, as compared to the wild-type allele, both y1-ww1 and y1-ww4 show greater susceptibility to the pathogenic fungus C. sublineolum. In fungal-inoculated wild-type seedlings, y1 and its target flavonoid structural genes are coordinately expressed. However, in y1-ww1 and y1-ww4 seedlings where y1 is not expressed, steady-state transcripts of its target genes could not be detected. Cosegregation analysis showed that the functional y1 gene is genetically linked with resistance to C. sublineolum. Overall results demonstrate that the accumulation of sorghum 3-deoxyanthocyanidin phytoalexins and resistance to C. sublineolum in sorghum require a functional y1 gene.
Figures



Similar articles
-
A sorghum MYB transcription factor induces 3-deoxyanthocyanidins and enhances resistance against leaf blights in maize.Molecules. 2015 Jan 30;20(2):2388-404. doi: 10.3390/molecules20022388. Molecules. 2015. PMID: 25647576 Free PMC article.
-
The wheat Lr34 multipathogen resistance gene confers resistance to anthracnose and rust in sorghum.Plant Biotechnol J. 2017 Nov;15(11):1387-1396. doi: 10.1111/pbi.12723. Epub 2017 Apr 20. Plant Biotechnol J. 2017. PMID: 28301718 Free PMC article.
-
Sorghum 3-Deoxyanthocyanidin Flavonoids Confer Resistance against Corn Leaf Aphid.J Chem Ecol. 2019 Jun;45(5-6):502-514. doi: 10.1007/s10886-019-01062-8. Epub 2019 Mar 26. J Chem Ecol. 2019. PMID: 30911880
-
Red card for pathogens: phytoalexins in sorghum and maize.Molecules. 2014 Jun 30;19(7):9114-33. doi: 10.3390/molecules19079114. Molecules. 2014. PMID: 24983861 Free PMC article. Review.
-
Temporal synthesis and radiolabelling of the sorghum 3-deoxyanthocyanidin phytoalexins and the anthocyanin, cyanidin 3-dimalonyl glucoside.New Phytol. 2000 Mar;145(3):457-469. doi: 10.1046/j.1469-8137.2000.00600.x. New Phytol. 2000. PMID: 33862912 Review.
Cited by
-
Loss of Pleiotropic Regulatory Functions in Tannin1, the Sorghum Ortholog of Arabidopsis Master Regulator TTG1.Plant Direct. 2025 Mar 12;9(3):e70055. doi: 10.1002/pld3.70055. eCollection 2025 Mar. Plant Direct. 2025. PMID: 40084038 Free PMC article.
-
RNAi of the sesquiterpene cyclase gene for phytoalexin production impairs pre- and post-invasive resistance to potato blight pathogens.Mol Plant Pathol. 2019 Jul;20(7):907-922. doi: 10.1111/mpp.12802. Epub 2019 Apr 16. Mol Plant Pathol. 2019. PMID: 30990946 Free PMC article.
-
Deciphering the role of phytoalexins in plant-microorganism interactions and human health.Molecules. 2014 Nov 5;19(11):18033-56. doi: 10.3390/molecules191118033. Molecules. 2014. PMID: 25379642 Free PMC article. Review.
-
Quantitative analysis of changes in the phosphoproteome of maize induced by the plant hormone salicylic acid.Sci Rep. 2015 Dec 11;5:18155. doi: 10.1038/srep18155. Sci Rep. 2015. PMID: 26659305 Free PMC article.
-
UV-induced reactive oxygen species and transcriptional control of 3-deoxyanthocyanidin biosynthesis in black sorghum pericarp.Front Plant Sci. 2024 Oct 7;15:1451215. doi: 10.3389/fpls.2024.1451215. eCollection 2024. Front Plant Sci. 2024. PMID: 39435026 Free PMC article.
References
-
- Aguero, M. E., A. Gevens and R. L. Nicholson, 2002. Interaction of Cochliobolus heterostrophus with phytoalexin inclusions in Sorghum bicolor. Physiol. Mol. Plant Pathol. 61 267–271.
-
- Basavaraju, P., N. P. Shetty, H. S. Shetty, E. de Neergaard and H. J. L. Jørgensen, 2009. Infection biology and defence responses in sorghum against Colletotrichum sublineolum. J. Appl. Microbiol. 107 404–415. - PubMed
-
- Berenbaum, M. R., and A. R. Zangerl, 1992. Genetics of secondary metabolism and herbivore resistance in plants, pp. 415–438 in Herbivores: Their Interactions With Secondary Plant Metabolites, edited by G. A. Rosenthal and M. R. Berenbaum. Academic Press, San Diego.
-
- Boddu, J., C. Svabek, R. Sekhon, A. Gevens, R. L. Nicholson et al., 2004. Expression of a putative flavonoid 3′-hydroxylase in sorghum mesocotyls synthesizing 3-deoxyanthocyanidin phytoalexins. Physiol. Mol. Plant Pathol. 65 101–113.
-
- Boddu, J., C. Svabek, F. Ibraheem, A. D. Jones and S. Chopra, 2005. Characterization of a deletion allele of a sorghum Myb gene, yellow seed1, showing loss of 3-deoxyflavonoids. Plant Sci. 169 542–552.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

